Rotavirus infection induces glycan availability to promote ileum-specific changes in the microbiome aiding rotavirus virulence
- PMID: 32404017
- PMCID: PMC7524290
- DOI: 10.1080/19490976.2020.1754714
Rotavirus infection induces glycan availability to promote ileum-specific changes in the microbiome aiding rotavirus virulence
Abstract
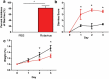
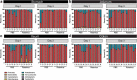
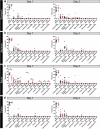
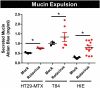
Multiple studies have identified changes within the gut microbiome in response to diarrheal-inducing bacterial pathogens. However, examination of the microbiome in response to viral pathogens remains understudied. Compounding this, many studies use fecal samples to assess microbiome composition; which may not accurately mirror changes within the small intestine, the primary site for most enteric virus infections. As a result, the functional significance of small intestinal microbiome shifts during infection is not well defined. To address these gaps, rotavirus-infected neonatal mice were examined for changes in bacterial community dynamics, host gene expression, and tissue recovery during infection. Profiling bacterial communities using 16S rRNA sequencing suggested significant and distinct changes in ileal communities in response to rotavirus infection, with no significant changes for other gastrointestinal (GI) compartments. At 1-d post-infection, we observed a loss in Lactobacillus species from the ileum, but an increase in Bacteroides and Akkermansia, both of which exhibit mucin-digesting capabilities. Concomitant with the bacterial community shifts, we observed a loss of mucin-filled goblet cells in the small intestine at d 1, with recovery occurring by d 3. Rotavirus infection of mucin-producing cell lines and human intestinal enteroids (HIEs) stimulated release of stored mucin granules, similar to in vivo findings. In vitro, incubation of mucins with Bacteroides or Akkermansia members resulted in significant glycan degradation, which altered the binding capacity of rotavirus in silico and in vitro. Taken together, these data suggest that the response to and recovery from rotavirus-diarrhea is unique between sub-compartments of the GI tract and may be influenced by mucin-degrading microbes.
Keywords: Akkermansia; Bacteroides; Muc2; Rotavirus; glycans; microbiome; mucus.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
