Frequency and spectrum of PIK3CA somatic mutations in breast cancer
- PMID: 32404150
- PMCID: PMC7222307
- DOI: 10.1186/s13058-020-01284-9
Frequency and spectrum of PIK3CA somatic mutations in breast cancer
Abstract
Purpose: The therascreen PIK3CA mutation assay and the alpha-specific PI3K inhibitor alpelisib are FDA-approved for identifying and treating patients with advanced PIK3CA-mutated (PIK3CAmut) breast cancer (BC). However, it is currently unknown to what extend this assay detects most PIK3CA mutations in BC. This information is critical as patients and clinicians are using this and other genomic assays to indicate alpelisib.
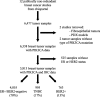
Methods: Data from 6338 patients with BC was explored across 10 publicly available studies. The primary objective was to evaluate the proportion and distribution of PIK3CA mutations in BC. Secondary objectives were (1) to evaluate in silico the spectrum of PIK3CA mutations in BC that would be captured by the therascreen panel; (2) to evaluate the proportion and distribution of PIK3CA mutations in hormone receptor-positive/HER2-negative (HR+/HER2-), HER2+, and triple-negative BC (TNBC); and (3) to explore the identification of PIK3CA mutations in a cohort of 48 HR+/HER2- advanced BC patients by the Guardant B360 circulating tumor DNA (ctDNA) assay.
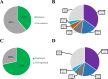
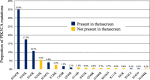
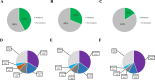
Results: Patients with PIK3CAmut tumors represented 35.7% (2261/6338). Five PIK3CA mutations comprised 73% of all PIK3CA mutations: H1047R (35%), E545K (17%), E542K (11%), N345K (6%), and H1047L (4%). Therascreen gene list would capture 72% of all PIK3CA mutations and 80% of patients with a known PIK3CAmut BC. Among patients with double PIK3CAmut tumors (12% of all PIK3CAmut), the therascreen panel would capture 78% as harboring 1 single PIK3CA mutation, 17% as PIK3CAmut undetected, and 5% as PIK3CA double-mut. PIK3CA mutation rates were lower in TNBC (16%) compared to HR+/HER2 (42%) and HER2+ (31%) BC; however, the distribution of the 4 main PIK3CA mutations across subtypes was similar. Finally, 28% of PIK3CA mutations identified in ctDNA in 48 patients with advanced HR+/HER2- BC were not part of the therascreen panel.
Conclusion: PIK3CA mutations in BC are heterogenous and ~ 20% of patients with a known PIK3CA mutation, and 95% with a known double PIK3CAmut tumor, would not be captured by the therascreen panel. Finally, the clinical utility of PIK3CA mutations not present in the therascreen companion diagnostic assay or identified by other sequencing-based assays needs further investigation.
Keywords: Alpelisib; Breast cancer; Companion diagnostic; Hotspot mutations; Mutations; PIK3CA; Therascreen; ctDNA.
Conflict of interest statement
AP reports that his institution received research funding from Nanostring Technologies, Roche, and Novartis. AP reports consulting and lecture fees from Nanostring Technologies, Roche, Novartis, Pfizer, Oncolytics Biotech, Amgen, Lilly, MSD, and PUMA.
Figures

References
-
- Mollon L, Aguilar A, Anderson E, et al. Abstract 1207: a systematic literature review of the prevalence of PIK3CA mutations and mutation hotspots in HR+/HER2- metastatic breast cancer. Cancer Res. 2018;78:1207.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

