Divergent domains of 28S ribosomal RNA gene: DNA barcodes for molecular classification and identification of mites
- PMID: 32404192
- PMCID: PMC7222323
- DOI: 10.1186/s13071-020-04124-z
Divergent domains of 28S ribosomal RNA gene: DNA barcodes for molecular classification and identification of mites
Abstract
Background: The morphological and molecular identification of mites is challenging due to the large number of species, the microscopic size of the organisms, diverse phenotypes of the same species, similar morphology of different species and a shortage of molecular data.
Methods: Nine medically important mite species belonging to six families, i.e. Demodex folliculorum, D. brevis, D. canis, D. caprae, Sarcoptes scabiei canis, Psoroptes cuniculi, Dermatophagoides farinae, Cheyletus malaccensis and Ornithonyssus bacoti, were collected and subjected to DNA barcoding. Sequences of cox1, 16S and 12S mtDNA, as well as ITS, 18S and 28S rDNA from mites were retrieved from GenBank and used as candidate genes. Sequence alignment and analysis identified 28S rDNA as the suitable target gene. Subsequently, universal primers of divergent domains were designed for molecular identification of 125 mite samples. Finally, the universality of the divergent domains with high identification efficiency was evaluated in Acari to screen DNA barcodes for mites.
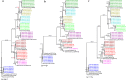
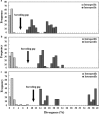
Results: Domains D5 (67.65%), D6 (62.71%) and D8 (77.59%) of the 28S rRNA gene had a significantly higher sequencing success rate, compared to domains D2 (19.20%), D3 (20.00%) and D7 (15.12%). The successful divergent domains all matched the closely-related species in GenBank with an identity of 74-100% and a coverage rate of 92-100%. Phylogenetic analysis also supported this result. Moreover, the three divergent domains had their own advantages. D5 had the lowest intraspecies divergence (0-1.26%), D6 had the maximum barcoding gap (10.54%) and the shortest sequence length (192-241 bp), and D8 had the longest indels (241 bp). Further universality analysis showed that the primers of the three divergent domains were suitable for identification across 225 species of 40 families in Acari.
Conclusions: This study confirmed that domains D5, D6 and D8 of 28S rDNA are universal DNA barcodes for molecular classification and identification of mites. 28S rDNA, as a powerful supplement for cox1 mtDNA 5'-end 648-bp fragment, recommended by the International Barcode of Life (IBOL), will provide great potential in molecular identification of mites in future studies because of its universality.
Keywords: DNA barcode; Divergent regions; Mites; Molecular identification; Universal primers; rDNA 28S.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
LSU rDNA D5 region: the DNA barcode for molecular classification and identification of Demodex.Genome. 2019 May;62(5):295-304. doi: 10.1139/gen-2018-0168. Epub 2019 Apr 18. Genome. 2019. PMID: 30998112
-
The nuclear 28S gene fragment D3 as species marker in oribatid mites (Acari, Oribatida) from German peatlands.Exp Appl Acarol. 2017 Mar;71(3):259-276. doi: 10.1007/s10493-017-0126-x. Epub 2017 Apr 12. Exp Appl Acarol. 2017. PMID: 28405837
-
Sequencing for complete rDNA sequences (18S, ITS1, 5.8S, ITS2, and 28S rDNA) of Demodex and phylogenetic analysis of Acari based on 18S and 28S rDNA.Parasitol Res. 2012 Nov;111(5):2109-14. doi: 10.1007/s00436-012-3058-8. Epub 2012 Aug 18. Parasitol Res. 2012. PMID: 22903416
-
DNA barcode markers applied to seafood authentication: an updated review.Crit Rev Food Sci Nutr. 2021;61(22):3904-3935. doi: 10.1080/10408398.2020.1811200. Epub 2020 Aug 25. Crit Rev Food Sci Nutr. 2021. PMID: 32838560 Review.
-
DNA-based methods for eriophyoid mite studies: review, critical aspects, prospects and challenges.Exp Appl Acarol. 2010 Jul;51(1-3):257-71. doi: 10.1007/s10493-009-9301-z. Epub 2009 Oct 14. Exp Appl Acarol. 2010. PMID: 19826904 Review.
Cited by
-
Novel Demodex detection method involving non-invasive sebum collection and next-generation sequencing.Postepy Dermatol Alergol. 2022 Apr;39(2):321-326. doi: 10.5114/ada.2021.106028. Epub 2021 Jul 16. Postepy Dermatol Alergol. 2022. PMID: 35645689 Free PMC article.
-
First detection of Ixodiphagus hookeri (Hymenoptera: Encyrtidae) in Ixodes ricinus ticks (Acari: Ixodidae) from multiple locations in Hungary.Sci Rep. 2023 Jan 28;13(1):1624. doi: 10.1038/s41598-023-28969-3. Sci Rep. 2023. PMID: 36709348 Free PMC article.
-
Molecular Identification and Phylogenetic Analysis of Laelapidae Mites (Acari: Mesostigmata).Animals (Basel). 2023 Jul 3;13(13):2185. doi: 10.3390/ani13132185. Animals (Basel). 2023. PMID: 37443981 Free PMC article.
-
Predation evaluation of the green lacewing, Chrysopa pallens on the pink tea mite pest, Acaphylla theae (Watt) (Acarina: Eriophyidae).Front Physiol. 2023 Dec 12;14:1307579. doi: 10.3389/fphys.2023.1307579. eCollection 2023. Front Physiol. 2023. PMID: 38152250 Free PMC article.
-
Wolbachia of phylogenetic supergroup K identified in oribatid mite Nothrus anauniensis (Acari: Oribatida: Nothridae).Exp Appl Acarol. 2024 Dec;93(4):803-815. doi: 10.1007/s10493-024-00961-0. Epub 2024 Sep 12. Exp Appl Acarol. 2024. PMID: 39266798
References
-
- Lima DB, Rezende-Puker D, Mendonca RS, Tixier MS, Gondim MGC, Melo JWS, et al. Molecular and morphological characterization of the predatory mite Amblyseius largoensis (Acari: Phytoseiidae): surprising similarity between an Asian and American populations. Exp Appl Acarol. 2018;76:287–310. doi: 10.1007/s10493-018-0308-1. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

