Gene Expression: the Key to Understanding HIV-1 Infection?
- PMID: 32404327
- PMCID: PMC7233484
- DOI: 10.1128/MMBR.00080-19
Gene Expression: the Key to Understanding HIV-1 Infection?
Abstract
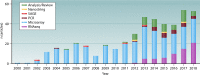
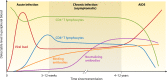
Gene expression profiling of the host response to HIV infection has promised to fill the gaps in our knowledge and provide new insights toward vaccine and cure. However, despite 20 years of research, the biggest questions remained unanswered. A literature review identified 62 studies examining gene expression dysregulation in samples from individuals living with HIV. Changes in gene expression were dependent on cell/tissue type, stage of infection, viremia, and treatment status. Some cell types, notably CD4+ T cells, exhibit upregulation of cell cycle, interferon-related, and apoptosis genes consistent with depletion. Others, including CD8+ T cells and natural killer cells, exhibit perturbed function in the absence of direct infection with HIV. Dysregulation is greatest during acute infection. Differences in study design and data reporting limit comparability of existing research and do not as yet provide a coherent overview of gene expression in HIV. This review outlines the extraordinarily complex host response to HIV and offers recommendations to realize the full potential of HIV host transcriptomics.
Keywords: gene expression; human immunodeficiency virus; transcriptional regulation; virus-host interactions.
Copyright © 2020 American Society for Microbiology.
Figures
References
-
- Ellis MJ, Perou CM. 2013. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov 3:27–34. doi:10.1158/2159-8290.CD-12-0462. - DOI - PMC - PubMed
-
- Costello JC, NCI DREAM Community, Heiser LM, Georgii E, Gönen M, Menden MP, Wang NJ, Bansal M, Ammad-Ud-Din M, Hintsanen P, Khan SA, Mpindi J-P, Kallioniemi O, Honkela A, Aittokallio T, Wennerberg K, NCI Dream Community, Collins JC, Gallahan D, Singer D, Saez-Rodriguez J, Kaski S, Gray JW, Stolovitzky G. 2014. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 32:1202–1212. doi:10.1038/nbt.2877. - DOI - PMC - PubMed
-
- Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff THM, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J, Shankar S, Parida SK, Kaufmann SHE, Walzl G, Aderem A, Hanekom WA. 2016. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387:2312–2322. doi:10.1016/S0140-6736(15)01316-1. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials