The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop
- PMID: 32404367
- PMCID: PMC7363137
- DOI: 10.1074/jbc.RA120.013590
The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop
Abstract
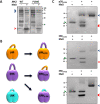
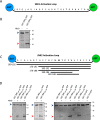
Microbial pathogens often target the host mitogen-activated protein kinase (MAPK) network to suppress host immune responses. We previously identified a bacterial type III secretion system effector, termed NleD, a metalloprotease that inactivates MAPKs by specifically cleaving their activation loop. Here, we show that NleDs form a growing family of virulence factors harbored by human and plant pathogens as well as insect symbionts. These NleDs disable specifically Jun N-terminal kinases (JNKs) and p38s that are required for host immune response, whereas extracellular signal-regulated kinase (ERK), which is essential for host cell viability, remains intact. We investigated the mechanism that makes ERK resistant to NleD cleavage. Biochemical and structural analyses revealed that NleD exclusively targets activation loops with high conformational flexibility. Accordingly, NleD cleaved the flexible loops of JNK and p38 but not the rigid loop of ERK. Our findings elucidate a compelling mechanism of native substrate proteolysis that is promoted by entropy-driven specificity. We propose that such entropy-based selectivity is a general attribute of proteolytic enzymes.
Keywords: Jun N-terminal kinase (JNK); NleD; bacterial pathogen; bacterial pathogenesis; crystallography; effector protein; extracellular signal-regulated kinase (ERK); metalloprotease; mitogen-activated protein kinase (MAPK); p38; protein structure; proteolysis; structural biology; type III secretion system (T3SS).
© 2020 Gur-Arie et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures



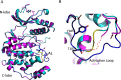
References
-
- Arbibe L., Kim D. W., Batsche E., Pedron T., Mateescu B., Muchardt C., Parsot C., and Sansonetti P. J. (2007) An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8, 47–56 10.1038/ni1423 - DOI - PubMed
-
- Stofega M. R., Yu C. L., Wu J., and Jove R. (1997) Activation of extracellular signal-regulated kinase (ERK) by mitogenic stimuli is repressed in v-Src-transformed cells. Cell Growth Differ. 8, 113–119 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

