Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts
- PMID: 32404529
- PMCID: PMC7375388
- DOI: 10.1128/JVI.00831-20
Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts
Abstract
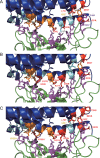
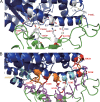
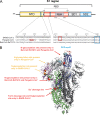
The emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), resulted in a pandemic. Here, we used X-ray structures of human ACE2 bound to the receptor-binding domain (RBD) of the spike protein (S) from SARS-CoV-2 to predict its binding to ACE2 proteins from different animals, including pets, farm animals, and putative intermediate hosts of SARS-CoV-2. Comparing the interaction sites of ACE2 proteins known to serve or not serve as receptors allows the definition of residues important for binding. From the 20 amino acids in ACE2 that contact S, up to 7 can be replaced and ACE2 can still function as the SARS-CoV-2 receptor. These variable amino acids are clustered at certain positions, mostly at the periphery of the binding site, while changes of the invariable residues prevent S binding or infection of the respective animal. Some ACE2 proteins even tolerate the loss or acquisition of N-glycosylation sites located near the S interface. Of note, pigs and dogs, which are not infected or are not effectively infected and have only a few changes in the binding site, exhibit relatively low levels of ACE2 in the respiratory tract. Comparison of the RBD of S of SARS-CoV-2 with that from bat coronavirus strain RaTG13 (Bat-CoV-RaTG13) and pangolin coronavirus (Pangolin-CoV) strain hCoV-19/pangolin/Guangdong/1/2019 revealed that the latter contains only one substitution, whereas Bat-CoV-RaTG13 exhibits five. However, ACE2 of pangolin exhibits seven changes relative to human ACE2, and a similar number of substitutions is present in ACE2 of bats, raccoon dogs, and civets, suggesting that SARS-CoV-2 may not be especially adapted to ACE2 of any of its putative intermediate hosts. These analyses provide new insight into the receptor usage and animal source/origin of SARS-CoV-2.IMPORTANCE SARS-CoV-2 is threatening people worldwide, and there are no drugs or vaccines available to mitigate its spread. The origin of the virus is still unclear, and whether pets and livestock can be infected and transmit SARS-CoV-2 are important and unknown scientific questions. Effective binding to the host receptor ACE2 is the first prerequisite for infection of cells and determines the host range. Our analysis provides a framework for the prediction of potential hosts of SARS-CoV-2. We found that ACE2 from species known to support SARS-CoV-2 infection tolerate many amino acid changes, indicating that the species barrier might be low. Exceptions are dogs and especially pigs, which revealed relatively low ACE2 expression levels in the respiratory tract. Monitoring of animals is necessary to prevent the generation of a new coronavirus reservoir. Finally, our analysis also showed that SARS-CoV-2 may not be specifically adapted to any of its putative intermediate hosts.
Keywords: SARS-CoV-2; angiotensin-converting enzyme 2; livestock; severe acute respiratory syndrome coronavirus 2.
Copyright © 2020 American Society for Microbiology.
Figures


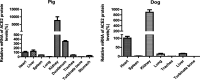

References
-
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Li M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JTK, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207. doi:10.1056/NEJMoa2001316. - DOI - PMC - PubMed
-
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. 2020. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323:1061–1069. doi:10.1001/jama.2020.1585. - DOI - PMC - PubMed
-
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. doi:10.1016/S0140-6736(20)30154-9. - DOI - PMC - PubMed
-
- Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC, Chen QX, Gao YW, Zhou HQ, Xiang H, Zheng HJ, Chern SW, Cheng F, Pan CM, Xuan H, Chen SJ, Luo HM, Zhou DH, Liu YF, He JF, Qin PZ, Li LH, Ren YQ, Liang WJ, Yu YD, Anderson L, Wang M, Xu RH, Wu XW, Zheng HY, Chen JD, Liang G, Gao Y, Liao M, Fang L, Jiang LY, Li H, Chen F, Di B, He LJ, Lin JY, Tong S, Kong X, Du L, Hao P, Tang H, Bernini A, Yu XJ, Spiga O, Guo ZM, Pan HY, He WZ, Manuguerra JC, Fontanet A, Danchin A, Niccolai N, Li YX, Wu CI, Zhao GP. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A 102:2430–2435. doi:10.1073/pnas.0409608102. - DOI - PMC - PubMed
-
- Hu B, Zeng L-P, Yang X-L, Ge X-Y, Zhang W, Li B, Xie J-Z, Shen X-R, Zhang Y-Z, Wang N, Luo D-S, Zheng X-S, Wang M-N, Daszak P, Wang L-F, Cui J, Shi Z-L. 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 13:e1006698. doi:10.1371/journal.ppat.1006698. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

