YAP1/TAZ drives ependymoma-like tumour formation in mice
- PMID: 32404936
- PMCID: PMC7220953
- DOI: 10.1038/s41467-020-16167-y
YAP1/TAZ drives ependymoma-like tumour formation in mice
Erratum in
-
Author Correction: YAP1/TAZ drives ependymoma-like tumour formation in mice.Nat Commun. 2020 Sep 28;11(1):4934. doi: 10.1038/s41467-020-18851-5. Nat Commun. 2020. PMID: 32985498 Free PMC article.
Abstract
YAP1 gene fusions have been observed in a subset of paediatric ependymomas. Here we show that, ectopic expression of active nuclear YAP1 (nlsYAP5SA) in ventricular zone neural progenitor cells using conditionally-induced NEX/NeuroD6-Cre is sufficient to drive brain tumour formation in mice. Neuronal differentiation is inhibited in the hippocampus. Deletion of YAP1's negative regulators LATS1 and LATS2 kinases in NEX-Cre lineage in double conditional knockout mice also generates similar tumours, which are rescued by deletion of YAP1 and its paralog TAZ. YAP1/TAZ-induced mouse tumours display molecular and ultrastructural characteristics of human ependymoma. RNA sequencing and quantitative proteomics of mouse tumours demonstrate similarities to YAP1-fusion induced supratentorial ependymoma. Finally, we find that transcriptional cofactor HOPX is upregulated in mouse models and in human YAP1-fusion induced ependymoma, supporting their similarity. Our results show that uncontrolled YAP1/TAZ activity in neuronal precursor cells leads to ependymoma-like tumours in mice.
Conflict of interest statement
The authors declare no competing interests.
Figures
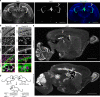
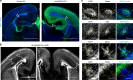
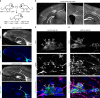
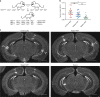


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

