Validation of SARS-CoV-2 detection across multiple specimen types
- PMID: 32405257
- PMCID: PMC7219399
- DOI: 10.1016/j.jcv.2020.104438
Validation of SARS-CoV-2 detection across multiple specimen types
Abstract
Background: Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has caused considerable disruption across the world, resulting in more than 235,000 deaths since December 2019. SARS-CoV-2 has a wide tropism and detection of the virus has been described in multiple specimen types, including various respiratory secretions, cerebrospinal fluid, and stool.
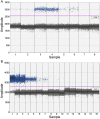
Objective: To evaluate the accuracy and sensitivity of a laboratory modified CDCbased SARS-CoV-2 N1 and N2 assay across a range of sample types. Study Design We compared the matrix effect on the analytical sensitivity of SARS-CoV-2 detection by qRT-PCR in nasal swabs collected in viral transport medium (VTM), bronchoalveolar lavage (BAL), sputum, plasma, cerebral spinal fluid (CSF), stool, VTM, phosphate buffered saline (PBS), and Hanks' Balanced Salt Solution (HBSS). Initial limits of detection (LoD) were subsequently narrowed to confirm an LoD for each specimen type and target gene.
Results: LoDs were established using a modified CDC-based laboratory developed test and ranged from a mean CT cut-off of 33.8-35.7 (10-20 copies/reaction) for the N1 gene target, and 34.0-36.2 (1-10 copies/reaction) for N2. Alternatives to VTM such as PBS and HBSS had comparable LoDs. The N2 gene target was found to be most sensitive in CSF.
Conclusion: A modified CDC-based laboratory developed test is able to detect SARSCoV- 2 accurately with similar sensitivity across all sample types tested.
Keywords: BAL; CSF; Coronavirus; SARS-CoV-2; Sensitivity; Specimen type.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures
References
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
-
- Yam W.C., Chan K.H., Poon L.L.M., Guan Y., Yuen K.Y., Seto W.H., Peiris J.S.M. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 2003;41:4521–4524. doi: 10.1128/JCM.41.10.4521-4524.2003. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

