The anti-tumor activity of pralatrexate (PDX) correlates with the expression of RFC and DHFR mRNA in preclinical models of multiple myeloma
- PMID: 32405334
- PMCID: PMC7210016
- DOI: 10.18632/oncotarget.27516
The anti-tumor activity of pralatrexate (PDX) correlates with the expression of RFC and DHFR mRNA in preclinical models of multiple myeloma
Abstract
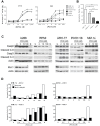
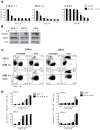
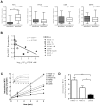
Multiple myeloma (MM) is the second most common hematologic malignancy. While major advances have been made in the disease, it is still incurable. Although antifolate-based drugs are not commonly used to treat myeloma, new generation analogs with distinct patterns of preclinical and clinical activity may offer an opportunity to identify new classes of potentially active drugs. Pralatrexate (PDX), which was approved for the treatment of relapsed or refractory peripheral T-cell lymphoma in 2009, may be one such drug. Pralatrexate exhibits a potency and pattern of activity distinct from its predecessors like methotrexate (MTX). We sought to understand the activity and mechanisms of resistance of multiple myeloma to these drugs, which could also offer potential strategies for selective use of the drug. We demonstrate that PDX and MTX both induce a significant decrease in cell viability in the low nanomolar range, with PDX exhibiting a more potent effect. We identified a series of myeloma cell lines exhibiting markedly different patterns of sensitivity to the drugs, with some lines frankly resistant, and others exquisitely sensitive. These differences were largely attributed to the basal RFC (Reduced Folate Carrier) mRNA expression levels. RFC mRNA expression correlated directly with rates of drug uptake, with the most sensitive lines exhibiting the most significant intracellular accumulation of pralatrexate. This mechanism explains the widely varying patterns of sensitivity and resistance to pralatrexate in multiple myeloma cell lines. These findings could have implications for this class of drugs and their role in the treatment of multiple myeloma.
Keywords: antifolate; biomarker; multiple myeloma; pralatrexate; reduced folate carrier (RFC).
Copyright: © 2020 Kinahan et al.
Conflict of interest statement
CONFLICTS OF INTEREST OAO receives research support from Spectrum Pharmaceuticals. HJC is an employee of the Multiple Myeloma Research Foundation, received commercial research support from Agenus Inc. and Genentech Roche, and compensation for consulting/advisory boards from Genentech Roche, Celgene, Bristol- Myers Squibb, and GlaxoSmithKline. All other authors declare no conflicts of interests.
Figures



References
-
- Laubach J, Garderet L, Mahindra A, Gahrton G, Caers J, Sezer O, Voorhees P, Leleu X, Johnsen HE, Streetly M, Jurczyszyn A, Ludwig H, Mellqvist UH, et al. . Management of relapsed multiple myeloma: recommendations of the International Myeloma Working Group. Leukemia. 2016; 30:1005–17. 10.1038/leu.2015.356. - DOI - PubMed
-
- Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, Middha S, Asmann Y, Schmidt J, Braggio E, Keats JJ, Fonseca R, Bergsagel PL, et al. . Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012; 120:1060–66. 10.1182/blood-2012-01-405977. - DOI - PMC - PubMed
-
- Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, Hinton JW, Li Y, Tubio JM, et al. . Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014; 5:2997. 10.1038/ncomms3997. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

