Characterization of the ZFX family of transcription factors that bind downstream of the start site of CpG island promoters
- PMID: 32406922
- PMCID: PMC7293018
- DOI: 10.1093/nar/gkaa384
Characterization of the ZFX family of transcription factors that bind downstream of the start site of CpG island promoters
Abstract
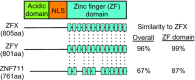
Our study focuses on a family of ubiquitously expressed human C2H2 zinc finger proteins comprised of ZFX, ZFY and ZNF711. Although their protein structure suggests that ZFX, ZFY and ZNF711 are transcriptional regulators, the mechanisms by which they influence transcription have not yet been elucidated. We used CRISPR-mediated deletion to create bi-allelic knockouts of ZFX and/or ZNF711 in female HEK293T cells (which naturally lack ZFY). We found that loss of either ZFX or ZNF711 reduced cell growth and that the double knockout cells have major defects in proliferation. RNA-seq analysis revealed that thousands of genes showed altered expression in the double knockout clones, suggesting that these TFs are critical regulators of the transcriptome. To gain insight into how these TFs regulate transcription, we created mutant ZFX proteins and analyzed them for DNA binding and transactivation capability. We found that zinc fingers 11-13 are necessary and sufficient for DNA binding and, in combination with the N terminal region, constitute a functional transactivator. Our functional analyses of the ZFX family provides important new insights into transcriptional regulation in human cells by members of the large, but under-studied family of C2H2 zinc finger proteins.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
ZFX acts as a transcriptional activator in multiple types of human tumors by binding downstream from transcription start sites at the majority of CpG island promoters.Genome Res. 2018 Mar 1;28(3):310-320. doi: 10.1101/gr.228809.117. Genome Res. 2018. PMID: 29429977 Free PMC article.
-
CpG islands in human ZFX and ZFY and mouse Zfx genes: sequence similarities and methylation differences.Genomics. 1995 Sep 20;29(2):353-63. doi: 10.1006/geno.1995.9994. Genomics. 1995. PMID: 8666382
-
Cloning and comparative analysis of the bovine, porcine, and equine sex chromosome genes ZFX and ZFY.Genome. 2004 Feb;47(1):74-83. doi: 10.1139/g03-099. Genome. 2004. PMID: 15060604
-
The mammalian zinc finger transcription factor Krüppel-like factor 3 (KLF3/BKLF).IUBMB Life. 2011 Feb;63(2):86-93. doi: 10.1002/iub.422. Epub 2011 Feb 24. IUBMB Life. 2011. PMID: 21360637 Review.
-
[Progress in the study of KRAB zinc finger protein].Yi Chuan. 2006 Nov;28(11):1451-6. doi: 10.1360/yc-006-1451. Yi Chuan. 2006. PMID: 17098717 Review. Chinese.
Cited by
-
High-throughput mRNA sequencing of human placenta shows sex differences across gestation.Placenta. 2024 May;150:8-21. doi: 10.1016/j.placenta.2024.03.005. Epub 2024 Mar 21. Placenta. 2024. PMID: 38537412 Free PMC article.
-
Large-scale transcriptomic analyses reveal downstream target genes of ZFY1 and ZFY2 transcription factors in male germ cells.Cell Death Differ. 2025 Aug 27. doi: 10.1038/s41418-025-01569-6. Online ahead of print. Cell Death Differ. 2025. PMID: 40866672
-
Stable and robust Xi and Y transcriptomes drive cell-type-specific autosomal and Xa responses in vivo and in vitro in four human cell types.Cell Genom. 2024 Sep 11;4(9):100628. doi: 10.1016/j.xgen.2024.100628. Epub 2024 Aug 6. Cell Genom. 2024. PMID: 39111319 Free PMC article.
-
Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma.Biomark Res. 2022 Jan 9;10(1):2. doi: 10.1186/s40364-021-00345-1. Biomark Res. 2022. PMID: 35000617 Free PMC article. Review.
-
Analysis of a Set of KDM5C Regulatory Genes Mutated in Neurodevelopmental Disorders Identifies Temporal Coexpression Brain Signatures.Genes (Basel). 2021 Jul 18;12(7):1088. doi: 10.3390/genes12071088. Genes (Basel). 2021. PMID: 34356104 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

