Application of Multiplex Bisulfite PCR-Ligase Detection Reaction-Real-Time Quantitative PCR Assay in Interrogating Bioinformatically Identified, Blood-Based Methylation Markers for Colorectal Cancer
- PMID: 32407802
- PMCID: PMC7338890
- DOI: 10.1016/j.jmoldx.2020.03.009
Application of Multiplex Bisulfite PCR-Ligase Detection Reaction-Real-Time Quantitative PCR Assay in Interrogating Bioinformatically Identified, Blood-Based Methylation Markers for Colorectal Cancer
Abstract
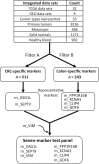
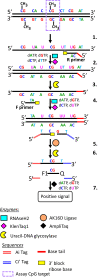
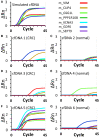
The analysis of CpG methylation in circulating tumor DNA fragments has emerged as a promising approach for the noninvasive early detection of solid tumors, including colorectal cancer (CRC). The most commonly employed assay involves bisulfite conversion of circulating tumor DNA, followed by targeted PCR, then real-time quantitative PCR (alias methylation-specific PCR). This report demonstrates the ability of a multiplex bisulfite PCR-ligase detection reaction-real-time quantitative PCR assay to detect seven methylated CpG markers (CRC or colon specific), in both simulated (approximately 30 copies of fragmented CRC cell line DNA mixed with approximately 3000 copies of fragmented peripheral blood DNA) and CRC patient-derived cell-free DNAs. This scalable assay is designed for multiplexing and incorporates steps for improved sensitivity and specificity, including the enrichment of methylated CpG fragments, ligase detection reaction, the incorporation of ribose bases in primers, and use of uracil DNA glycosylase. Six of the seven CpG markers (located in promoter regions of PPP1R16B, KCNA3, CLIP4, GDF6, SEPT9, and GSG1L) were identified through integrated analyses of genome-wide methylation data sets for 31 different types of cancer. These markers were mapped to CpG sites at the promoter region of VIM; VIM and SEPT9 are established epigenetic markers of CRC. Additional bioinformatics analyses show that the methylation at these CpG sites negatively correlates with the transcription of their corresponding genes.
Copyright © 2020 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Siegel R.L., Miller K.D., Fedewa S.A., Ahnen D.J., Meester R.G.S., Barzi A., Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. - PubMed
-
- Rex D.K., Boland C.R., Dominitz J.A., Giardiello F.M., Johnson D.A., Kaltenbach T., Levin T.R., Lieberman D., Robertson D.J. Colorectal cancer screening: recommendations for physicians and patients from the U.S. Multi-Society task force on colorectal cancer. Gastroenterology. 2017;153:307–323. - PubMed
-
- Beydoun H.A., Beydoun M.A. Predictors of colorectal cancer screening behaviors among average-risk older adults in the United States. Cancer Causes Control. 2008;19:339–359. - PubMed
-
- Das V., Kalita J., Pal M. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed Pharmacother. 2016;87:8–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

