Analysis of shared common genetic risk between amyotrophic lateral sclerosis and epilepsy
- PMID: 32409253
- PMCID: PMC7818383
- DOI: 10.1016/j.neurobiolaging.2020.04.011
Analysis of shared common genetic risk between amyotrophic lateral sclerosis and epilepsy
Abstract
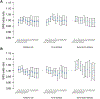
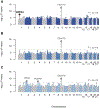
Because hyper-excitability has been shown to be a shared pathophysiological mechanism, we used the latest and largest genome-wide studies in amyotrophic lateral sclerosis (n = 36,052) and epilepsy (n = 38,349) to determine genetic overlap between these conditions. First, we showed no significant genetic correlation, also when binned on minor allele frequency. Second, we confirmed the absence of polygenic overlap using genomic risk score analysis. Finally, we did not identify pleiotropic variants in meta-analyses of the 2 diseases. Our findings indicate that amyotrophic lateral sclerosis and epilepsy do not share common genetic risk, showing that hyper-excitability in both disorders has distinct origins.
Keywords: ALS; Epilepsy; Genetic correlation.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
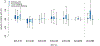
References
-
- van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, van der Spek RA, Võsa U, de Jong S, Robinson MR, Yang J, Fogh I, van Doormaal PT, Tazelaar GH, 2016. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet 48, 1043–1048. - PMC - PubMed

