Convergent Loss of an EDS1/PAD4 Signaling Pathway in Several Plant Lineages Reveals Coevolved Components of Plant Immunity and Drought Response
- PMID: 32409319
- PMCID: PMC7346574
- DOI: 10.1105/tpc.19.00903
Convergent Loss of an EDS1/PAD4 Signaling Pathway in Several Plant Lineages Reveals Coevolved Components of Plant Immunity and Drought Response
Abstract
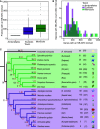
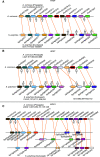
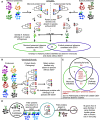
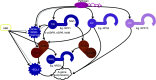
Plant innate immunity relies on nucleotide binding leucine-rich repeat receptors (NLRs) that recognize pathogen-derived molecules and activate downstream signaling pathways. We analyzed the variation in NLR gene copy number and identified plants with a low number of NLR genes relative to sister species. We specifically focused on four plants from two distinct lineages, one monocot lineage (Alismatales) and one eudicot lineage (Lentibulariaceae). In these lineages, the loss of NLR genes coincides with loss of the well-known downstream immune signaling complex ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1)/PHYTOALEXIN DEFICIENT 4 (PAD4). We expanded our analysis across whole proteomes and found that other characterized immune genes were absent only in Lentibulariaceae and Alismatales. Additionally, we identified genes of unknown function that were convergently lost together with EDS1/PAD4 in five plant species. Gene expression analyses in Arabidopsis (Arabidopsis thaliana) and Oryza sativa revealed that several homologs of the candidates are differentially expressed during pathogen infection, drought, and abscisic acid treatment. Our analysis provides evolutionary evidence for the rewiring of plant immunity in some plant lineages, as well as the coevolution of the EDS1/PAD4 pathway and drought responses.
© 2020 American Society of Plant Biologists. All rights reserved.
Figures




Comment in
-
ASTREL Projection: Comparative Phylogenomics Uncovers Novel Genes Coeliminated with the EDS1 Immune Pathway.Plant Cell. 2020 Jul;32(7):2067-2068. doi: 10.1105/tpc.20.00402. Epub 2020 May 28. Plant Cell. 2020. PMID: 32467110 Free PMC article. No abstract available.
References
-
- Baggs E., Dagdas G., Krasileva K.V.(2017). NLR diversity, helpers and integrated domains: Making sense of the NLR IDentity. Curr. Opin. Plant Biol. 38: 59–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/CSP17270/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/T/000PR9818/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/T/000PR9783 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011216/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

