A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation
- PMID: 32409580
- PMCID: PMC7363119
- DOI: 10.1074/jbc.RA120.013028
A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation
Abstract
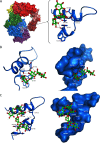
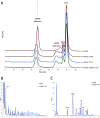
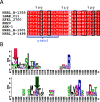
Microbial α-glucans produced by GH70 (glycoside hydrolase family 70) glucansucrases are gaining importance because of the mild conditions for their synthesis from sucrose, their biodegradability, and their current and anticipated applications that largely depend on their molar mass. Focusing on the alternansucrase (ASR) from Leuconostoc citreum NRRL B-1355, a well-known glucansucrase catalyzing the synthesis of both high- and low-molar-mass alternans, we searched for structural traits in ASR that could be involved in the control of alternan elongation. The resolution of five crystal structures of a truncated ASR version (ASRΔ2) in complex with different gluco-oligosaccharides pinpointed key residues in binding sites located in the A and V domains of ASR. Biochemical characterization of three single mutants and three double mutants targeting the sugar-binding pockets identified in domain V revealed an involvement of this domain in alternan binding and elongation. More strikingly, we found an oligosaccharide-binding site at the surface of domain A, distant from the catalytic site and not previously identified in other glucansucrases. We named this site surface-binding site (SBS) A1. Among the residues lining the SBS-A1 site, two (Gln700 and Tyr717) promoted alternan elongation. Their substitution to alanine decreased high-molar-mass alternan yield by a third, without significantly impacting enzyme stability or specificity. We propose that the SBS-A1 site is unique to alternansucrase and appears to be designed to bind alternating structures, acting as a mediator between the catalytic site and the sugar-binding pockets of domain V and contributing to a processive elongation of alternan chains.
Keywords: GH70; alternan; alternansucrase; biosourced; biotechnology; carbohydrate structure; dextran; enzyme catalysis; enzyme mechanism; glucansucrase; green chemistry; polymerase; polysaccharide; processivity; sugar-binding pockets; surface-binding site.
© 2020 Molina et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures





Similar articles
-
Overview of the glucansucrase equipment of Leuconostoc citreum LBAE-E16 and LBAE-C11, two strains isolated from sourdough.FEMS Microbiol Lett. 2015 Jan;362(1):1-8. doi: 10.1093/femsle/fnu024. Epub 2014 Dec 4. FEMS Microbiol Lett. 2015. PMID: 25790502
-
Unravelling Regioselectivity of Leuconostoc citreum ABK-1 Alternansucrase by Acceptor Site Engineering.Int J Mol Sci. 2021 Mar 22;22(6):3229. doi: 10.3390/ijms22063229. Int J Mol Sci. 2021. PMID: 33810084 Free PMC article.
-
An α-1,6-and α-1,3-linked glucan produced by Leuconostoc citreum ABK-1 alternansucrase with nanoparticle and film-forming properties.Sci Rep. 2018 May 29;8(1):8340. doi: 10.1038/s41598-018-26721-w. Sci Rep. 2018. PMID: 29844508 Free PMC article.
-
Alternansucrase as a key enabling tool of biotransformation from molecular features to applications: A review.Int J Biol Macromol. 2024 Nov;279(Pt 1):135096. doi: 10.1016/j.ijbiomac.2024.135096. Epub 2024 Aug 28. Int J Biol Macromol. 2024. PMID: 39214198 Review.
-
Extracellular polysaccharides produced by bacteria of the Leuconostoc genus.World J Microbiol Biotechnol. 2020 Sep 29;36(11):161. doi: 10.1007/s11274-020-02937-9. World J Microbiol Biotechnol. 2020. PMID: 32989599 Review.
Cited by
-
Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure-Function Relationship Studies and Engineering.Microorganisms. 2021 Jul 28;9(8):1607. doi: 10.3390/microorganisms9081607. Microorganisms. 2021. PMID: 34442685 Free PMC article. Review.
-
Architecture, Function, Regulation, and Evolution of α-Glucans Metabolic Enzymes in Prokaryotes.Chem Rev. 2024 Apr 24;124(8):4863-4934. doi: 10.1021/acs.chemrev.3c00811. Epub 2024 Apr 12. Chem Rev. 2024. PMID: 38606812 Free PMC article. Review.
-
Unprecedented Diversity of the Glycoside Hydrolase Family 70: A Comprehensive Analysis of Sequence, Structure, and Function.J Agric Food Chem. 2024 Jul 31;72(30):16911-16929. doi: 10.1021/acs.jafc.4c04807. Epub 2024 Jul 18. J Agric Food Chem. 2024. PMID: 39025827 Free PMC article.
References
-
- Leemhuis H., Pijning T., Dobruchowska J. M., van Leeuwen S. S., Kralj S., Dijkstra B. W., and Dijkhuizen L. (2013) Glucansucrases: three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 163, 250–272 10.1016/j.jbiotec.2012.06.037 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

