The unfoldase ClpC1 of Mycobacterium tuberculosis regulates the expression of a distinct subset of proteins having intrinsically disordered termini
- PMID: 32409584
- PMCID: PMC7363115
- DOI: 10.1074/jbc.RA120.013456
The unfoldase ClpC1 of Mycobacterium tuberculosis regulates the expression of a distinct subset of proteins having intrinsically disordered termini
Abstract
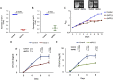
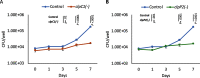
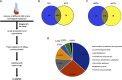
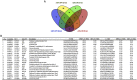
The human pathogen Mycobacterium tuberculosis (Mtb) harbors a well-orchestrated Clp (caseinolytic protease) proteolytic machinery consisting of two oligomeric segments, a barrel-shaped heterotetradecameric protease core comprising the ClpP1 and ClpP2 subunits, and hexameric ring-like ATP-dependent unfoldases composed of ClpX or ClpC1. The roles of the ClpP1P2 protease subunits are well-established in Mtb, but the potential roles of the associated unfoldases, such as ClpC1, remain elusive. Using a CRISPR interference-mediated gene silencing approach, here we demonstrate that clpC1 is indispensable for the extracellular growth of Mtb and for its survival in macrophages. The results from isobaric tags for relative and absolute quantitation-based quantitative proteomic experiments with clpC1- and clpP2-depleted Mtb cells suggested that the ClpC1P1P2 complex critically maintains the homeostasis of various growth-essential proteins in Mtb, several of which contain intrinsically disordered regions at their termini. We show that the Clp machinery regulates dosage-sensitive proteins such as the small heat shock protein Hsp20, which exists in a dodecameric conformation. Further, we observed that Hsp20 is poorly expressed in WT Mtb and that its expression is greatly induced upon depletion of clpC1 or clpP2 Remarkably, high Hsp20 protein levels were detected in the clpC1(-) or clpP2(-) knockdown strains but not in the parental bacteria, despite significant induction of hsp20 transcripts. In summary, the cellular levels of oligomeric proteins such as Hsp20 are maintained post-translationally through their recognition, disassembly, and degradation by ClpC1, which requires disordered ends in its protein substrates.
Keywords: ATPase; ClpC1 unfoldase; Hsp20; Mycobacterium tuberculosis; caseinolytic protease; heat shock protein (HSP); intrinsically disordered region; protein degradation; proteolysis; tuberculosis.
© 2020 Lunge et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

