Evidence of increasing diversification of emerging Severe Acute Respiratory Syndrome Coronavirus 2 strains
- PMID: 32410229
- PMCID: PMC7273070
- DOI: 10.1002/jmv.26018
Evidence of increasing diversification of emerging Severe Acute Respiratory Syndrome Coronavirus 2 strains
Abstract
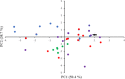
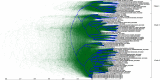
On 30th January 2020, an outbreak of atypical pneumonia caused by a novel betacoronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared a public health emergency of international concern by the World Health Organization. For this reason, a detailed evolutionary analysis of SARS-CoV-2 strains currently circulating in different geographic regions of the world was performed. A compositional analysis as well as a Bayesian coalescent analysis of complete genome sequences of SARS-CoV-2 strains recently isolated in Europe, North America, South America, and Asia was performed. The results of these studies revealed a diversification of SARS-CoV-2 strains in three different genetic clades. Co-circulation of different clades in different countries, as well as different genetic lineages within different clades were observed. The time of the most recent common ancestor was established to be around 1st November 2019. A mean rate of evolution of 6.57 × 10-4 substitutions per site per year was found. A significant migration rate per genetic lineage per year from Europe to South America was also observed. The results of these studies revealed an increasing diversification of SARS-CoV-2 strains. High evolutionary rates and fast population growth characterizes the population dynamics of SARS-CoV-2 strains.
Keywords: SARS-CoV-2; coalescent; coronavirus; evolution.
© 2020 Wiley Periodicals LLC.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

