Dynamic distribution of gut microbiota in meat rabbits at different growth stages and relationship with average daily gain (ADG)
- PMID: 32410629
- PMCID: PMC7227296
- DOI: 10.1186/s12866-020-01797-5
Dynamic distribution of gut microbiota in meat rabbits at different growth stages and relationship with average daily gain (ADG)
Abstract
Background: The mammalian intestinal tract harbors diverse and dynamic microbial communities that play pivotal roles in host health, metabolism, immunity, and development. Average daily gain (ADG) is an important growth trait in meat rabbit industry. The effects of gut microbiota on ADG in meat rabbits are still unknown.
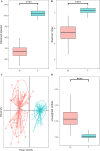
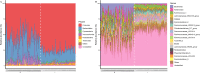
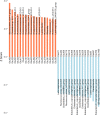
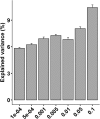
Results: In this study, we investigated the dynamic distribution of gut microbiota in commercial Ira rabbits from weaning to finishing and uncover the relationship between the microbiota and average daily gain (ADG) via 16S rRNA gene sequencing. The results indicated that the richness and diversity of gut microbiota significantly increased with age. Gut microbial structure was less variable among finishing rabbits than among weaning rabbits. The relative abundances of the dominant phyla Firmicutes, Bacteroidetes, Verrucomicrobia and Cyanobacteria, and the 15 predominant genera significantly varied with age. Metagenomic prediction analysis showed that both KOs and KEGG pathways related to the metabolism of monosaccharides and vitamins were enriched in the weaning rabbits, while those related to the metabolism of amino acids and polysaccharides were more abundant in the finishing rabbits. We identified 34 OTUs, 125 KOs, and 25 KEGG pathways that were significantly associated with ADG. OTUs annotation suggested that butyrate producing bacteria belong to the family Ruminococcaceae and Bacteroidales_S24-7_group were positively associated with ADG. Conversely, Eubacterium_coprostanoligenes_group, Christensenellaceae_R-7_group, and opportunistic pathogens were negatively associated with ADG. Both KOs and KEGG pathways correlated with the metabolism of vitamins, basic amino acids, and short chain fatty acids (SCFAs) showed positive correlations with ADG, while those correlated with aromatic amino acids metabolism and immune response exhibited negative correlations with ADG. In addition, our results suggested that 10.42% of the variation in weaning weight could be explained by the gut microbiome.
Conclusions: Our findings give a glimpse into the dynamic shifts in gut microbiota of meat rabbits and provide a theoretical basis for gut microbiota modulation to improve ADG in the meat rabbit industry.
Keywords: 16S rRNA; ADG; Dynamic distribution; Gut microbiota; Ira rabbits.
Conflict of interest statement
All authors declare that they have no competing interests.
Figures


Similar articles
-
Faecal microbiota and functional capacity associated with weaning weight in meat rabbits.Microb Biotechnol. 2019 Nov;12(6):1441-1452. doi: 10.1111/1751-7915.13485. Epub 2019 Oct 1. Microb Biotechnol. 2019. PMID: 31571427 Free PMC article.
-
Effect of host breeds on gut microbiome and serum metabolome in meat rabbits.BMC Vet Res. 2021 Jan 7;17(1):24. doi: 10.1186/s12917-020-02732-6. BMC Vet Res. 2021. PMID: 33413361 Free PMC article.
-
Effects of Gut Microbiome and Short-Chain Fatty Acids (SCFAs) on Finishing Weight of Meat Rabbits.Front Microbiol. 2020 Aug 11;11:1835. doi: 10.3389/fmicb.2020.01835. eCollection 2020. Front Microbiol. 2020. PMID: 32849435 Free PMC article.
-
Gut Microbiota Profile in Patients with Type 1 Diabetes Based on 16S rRNA Gene Sequencing: A Systematic Review.Dis Markers. 2020 Aug 27;2020:3936247. doi: 10.1155/2020/3936247. eCollection 2020. Dis Markers. 2020. PMID: 32908614 Free PMC article.
-
Structural diversity, functional aspects and future therapeutic applications of human gut microbiome.Arch Microbiol. 2021 Nov;203(9):5281-5308. doi: 10.1007/s00203-021-02516-y. Epub 2021 Aug 17. Arch Microbiol. 2021. PMID: 34405262 Free PMC article. Review.
Cited by
-
Genetic assessment of litter size, body weight, carcass traits and gene expression profiles in exotic and indigenous rabbit breeds: a study on New Zealand White, Californian, and Gabali rabbits in Egypt.Trop Anim Health Prod. 2024 Aug 22;56(7):244. doi: 10.1007/s11250-024-04082-z. Trop Anim Health Prod. 2024. PMID: 39172291 Free PMC article.
-
Genetic parameters of growth traits and carcass weight of New Zealand white rabbits in a tropical dry forest area.J Adv Vet Anim Res. 2021 Sep 20;8(3):471-478. doi: 10.5455/javar.2021.h536. eCollection 2021 Sep. J Adv Vet Anim Res. 2021. PMID: 34722746 Free PMC article.
-
The Effects of Temperature and Humidity Index on Growth Performance, Colon Microbiota, and Serum Metabolome of Ira Rabbits.Animals (Basel). 2023 Jun 13;13(12):1971. doi: 10.3390/ani13121971. Animals (Basel). 2023. PMID: 37370481 Free PMC article.
-
Multi-Omics Analysis Revealed the Molecular Mechanisms Affecting Average Daily Gain in Cattle.Int J Mol Sci. 2025 Mar 6;26(5):2343. doi: 10.3390/ijms26052343. Int J Mol Sci. 2025. PMID: 40076961 Free PMC article.
-
Association between body weight and distal gut microbes in Hainan black goats at weaning age.Front Microbiol. 2022 Sep 16;13:951473. doi: 10.3389/fmicb.2022.951473. eCollection 2022. Front Microbiol. 2022. PMID: 36187995 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

