Identification of novel, clinically correlated autoantigens in the monogenic autoimmune syndrome APS1 by proteome-wide PhIP-Seq
- PMID: 32410729
- PMCID: PMC7228772
- DOI: 10.7554/eLife.55053
Identification of novel, clinically correlated autoantigens in the monogenic autoimmune syndrome APS1 by proteome-wide PhIP-Seq
Abstract
The identification of autoantigens remains a critical challenge for understanding and treating autoimmune diseases. Autoimmune polyendocrine syndrome type 1 (APS1), a rare monogenic form of autoimmunity, presents as widespread autoimmunity with T and B cell responses to multiple organs. Importantly, autoantibody discovery in APS1 can illuminate fundamental disease pathogenesis, and many of the antigens found in APS1 extend to more common autoimmune diseases. Here, we performed proteome-wide programmable phage-display (PhIP-Seq) on sera from a cohort of people with APS1 and discovered multiple common antibody targets. These novel APS1 autoantigens exhibit tissue-restricted expression, including expression in enteroendocrine cells, pineal gland, and dental enamel. Using detailed clinical phenotyping, we find novel associations between autoantibodies and organ-restricted autoimmunity, including a link between anti-KHDC3L autoantibodies and premature ovarian insufficiency, and between anti-RFX6 autoantibodies and diarrheal-type intestinal dysfunction. Our study highlights the utility of PhIP-Seq for extensively interrogating antigenic repertoires in human autoimmunity and the importance of antigen discovery for improved understanding of disease mechanisms.
Keywords: APECED; PhIP-Seq; autoantigens; autoimmunity; enteroendocrine cells; human; human biology; immunology; inflammation; medicine; ovarian insufficiency.
Plain language summary
The immune system uses antibodies to fight microbes that cause disease. White blood cells pump antibodies into the bloodstream, and these antibodies latch onto bacteria and viruses, targeting them for destruction. But sometimes, the immune system gets it wrong. In autoimmune diseases, white blood cells mistakenly make antibodies that target the body's own tissues. Detecting these 'autoantibodies' in the blood can help doctors to diagnose autoimmune diseases. But the identities and targets of many autoantibodies remain unknown. In one rare disease, called autoimmune polyendocrine syndrome type 1 (APS-1), a faulty gene makes the immune system much more likely to make autoantibodies. People with this disease can develop an autoimmune response against many different healthy organs. Although APS-1 is rare, some of the autoantibodies made by individuals with the disease are the same as the ones in more common autoimmune diseases, like type 1 diabetes. Therefore, investigating the other autoantibodies produced by individuals with APS-1 could reveal the autoantibodies driving other autoimmune diseases. Autoantibodies bind to specific regions of healthy proteins, and one way to identify them is to use hundreds of thousands of tiny viruses in a technique called proteome-wide programmable phage-display, or PhIP-Seq. Each phage carries one type of protein segment. When mixed with blood serum from a patient, the autoantibodies stick to the phages that carry the target proteins for that autoantibody. These complexes can be isolated using biochemical techniques. Sequencing the genes of these phages then reveals the identity of the autoantibodies’ targets. Using this technique, Vazquez et al successfully pulled 23 known autoantibodies from the serum of patients with APS-1. Then, experiments to search for new targets began. This revealed many new autoantibodies, targeting proteins found only in specific tissues. They included one that targets a protein found on cells in the gut, and another that targets a protein found on egg cells in the ovaries. Matching the PhIP-Seq data to patient symptoms confirmed that these new antibodies correlate with the features of specific autoimmune diseases. For example, patients with antibodies that targeted the gut protein were more likely to have gut symptoms, while patients with antibodies that targeted the egg cell protein were more likely to have problems with their ovaries. Further investigations using PhIP-Seq could reveal the identities of even more autoantibodies. This might pave the way for new antibody tests to diagnose autoimmune diseases and identify tissues at risk of damage. This could be useful not only for people with APS-1, but also for more common autoimmune diseases that target the same organs.
Conflict of interest statement
SV JD, MSA, and SEV have a provisional patent on clinical application of autoantigens described in this study. EF, DS, SS, BM, CM, ZQ, AC, MC, MG, ML No competing interests declared, JD JD is a scientific advisory board member of Allen and Company. JD, MSA, and SEV have a provisional patent on clinical application of autoantigens described in this study. MA MSA owns stock in Merck and Medtronic. JD, MSA, and SEV have a provisional patent on clinical application of autoantigens described in this study.
Figures

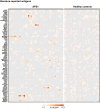

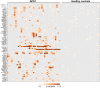

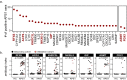
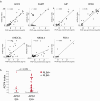

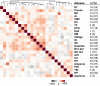
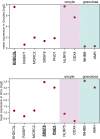
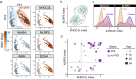
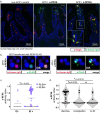


References
-
- Aaltonen J, Björses P, Perheentupa J, Horelli–Kuitunen N, Palotie A, Peltonen L, Lee YS, Francis F, Henning S, Thiel C, Leharach H, Yaspo M–L, Yaspo M, Finnish-German APECED Consortium An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nature Genetics. 1997;17:399–403. doi: 10.1038/ng1297-399. - DOI - PubMed
-
- Adriaenssens AE, Biggs EK, Darwish T, Tadross J, Sukthankar T, Girish M, Polex-Wolf J, Lam BY, Zvetkova I, Pan W, Chiarugi D, Yeo GSH, Blouet C, Gribble FM, Reimann F. Glucose-Dependent insulinotropic polypeptide Receptor-Expressing cells in the hypothalamus regulate food intake. Cell Metabolism. 2019;30:987–996. doi: 10.1016/j.cmet.2019.07.013. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Dryad/10.7272/Q66H4FM2
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

