Identification and validation of hub microRNAs dysregulated in esophageal squamous cell carcinoma
- PMID: 32412911
- PMCID: PMC7288914
- DOI: 10.18632/aging.103245
Identification and validation of hub microRNAs dysregulated in esophageal squamous cell carcinoma
Abstract
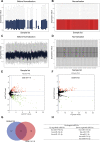
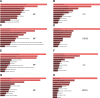
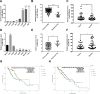
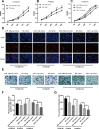
Esophageal squamous cell carcinoma (ESCC) is one of the deadliest cancers worldwide, and its morbidity is exacerbated by the lack of early symptoms. Bioinformatics analyses enable discovery of differentially expressed genes and non-protein-coding RNAs of potential prognostic and/or therapeutic relevance in ESCC and other cancers. Using bioinformatics tools, we searched for dysregulated miRNAs in two ESCC microarray datasets from the Gene Expression Omnibus (GEO) database. After identification of three upregulated and five downregulated miRNAs shared between databases, protein-protein interaction (PPI) network analysis was used to identify the top 10 hub-gene targets. Thereafter, a miRNA-gene interaction network predicted that most hub genes are regulated by miR-196a-5p and miR-1-3p, which are respectively upregulated and downregulated in ESCC. Functional enrichment analyses in the GO and KEGG databases indicated the potential involvement of these miRNAs in tumorigenesis-related processes and pathways, while both differential expression and correlation with T stage were demonstrated for each miRNA in a cohort of ESCC patients. Overexpression showed that miR-196a-5p increased, whereas miR-1-3p attenuated, proliferation and invasion in human ESCC cell lines grown in vitro. These findings suggest miR-196a-5p and miR-1-3p jointly contribute to ESCC tumorigenesis and are potential targets for diagnosis and treatment.
Keywords: bioinformatics; esophageal squamous cell carcinoma; hub miRNAs; tumorigenesis.
Conflict of interest statement
Figures



Similar articles
-
Identification of crucial miRNAs and genes in esophageal squamous cell carcinoma by miRNA-mRNA integrated analysis.Medicine (Baltimore). 2019 Jul;98(27):e16269. doi: 10.1097/MD.0000000000016269. Medicine (Baltimore). 2019. PMID: 31277149 Free PMC article.
-
Identification of Differentially Expressed Genes and miRNAs Associated with Esophageal Squamous Cell Carcinoma by Integrated Analysis of Microarray Data.Biomed Res Int. 2020 Jul 1;2020:1980921. doi: 10.1155/2020/1980921. eCollection 2020. Biomed Res Int. 2020. PMID: 32714975 Free PMC article.
-
Identification of invasion-metastasis-associated microRNAs in hepatocellular carcinoma based on bioinformatic analysis and experimental validation.J Transl Med. 2018 Sep 29;16(1):266. doi: 10.1186/s12967-018-1639-8. J Transl Med. 2018. PMID: 30268144 Free PMC article.
-
Common gene signatures and key pathways in hypopharyngeal and esophageal squamous cell carcinoma: Evidence from bioinformatic analysis.Medicine (Baltimore). 2020 Oct 16;99(42):e22434. doi: 10.1097/MD.0000000000022434. Medicine (Baltimore). 2020. PMID: 33080677 Free PMC article.
-
MicroRNAs in esophageal squamous cell carcinoma: Application in prognosis, diagnosis, and drug delivery.Pathol Res Pract. 2022 Dec;240:154196. doi: 10.1016/j.prp.2022.154196. Epub 2022 Nov 2. Pathol Res Pract. 2022. PMID: 36356334 Review.
Cited by
-
Identification of characteristic genes and construction of regulatory network in gallbladder carcinoma.BMC Med Genomics. 2023 Oct 11;16(1):240. doi: 10.1186/s12920-023-01663-z. BMC Med Genomics. 2023. PMID: 37821907 Free PMC article.
-
Carnosic Acid Induces Antiproliferation and Anti-Metastatic Property of Esophageal Cancer Cells via MAPK Signaling Pathways.J Oncol. 2021 Nov 16;2021:4451533. doi: 10.1155/2021/4451533. eCollection 2021. J Oncol. 2021. PMID: 34824582 Free PMC article.
-
Novel miRNA markers and their mechanism of esophageal squamous cell carcinoma (ESCC) based on TCGA.Sci Rep. 2024 Nov 8;14(1):27261. doi: 10.1038/s41598-024-76321-0. Sci Rep. 2024. PMID: 39516222 Free PMC article.
-
Long noncoding RNA steroid receptor RNA activator 1 inhibits proliferation and glycolysis of esophageal squamous cell carcinoma.World J Gastrointest Oncol. 2024 Oct 15;16(10):4194-4208. doi: 10.4251/wjgo.v16.i10.4194. World J Gastrointest Oncol. 2024. PMID: 39473951 Free PMC article.
-
Comprehensive analyses reveal the carcinogenic and immunological roles of ANLN in human cancers.Cancer Cell Int. 2022 May 14;22(1):188. doi: 10.1186/s12935-022-02610-1. Cancer Cell Int. 2022. PMID: 35568883 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

