Genomic Analysis of European Drosophila melanogaster Populations Reveals Longitudinal Structure, Continent-Wide Selection, and Previously Unknown DNA Viruses
- PMID: 32413142
- PMCID: PMC7475034
- DOI: 10.1093/molbev/msaa120
Genomic Analysis of European Drosophila melanogaster Populations Reveals Longitudinal Structure, Continent-Wide Selection, and Previously Unknown DNA Viruses
Abstract
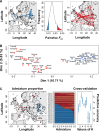
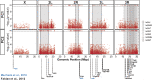
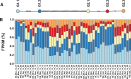
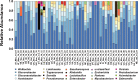
Genetic variation is the fuel of evolution, with standing genetic variation especially important for short-term evolution and local adaptation. To date, studies of spatiotemporal patterns of genetic variation in natural populations have been challenging, as comprehensive sampling is logistically difficult, and sequencing of entire populations costly. Here, we address these issues using a collaborative approach, sequencing 48 pooled population samples from 32 locations, and perform the first continent-wide genomic analysis of genetic variation in European Drosophila melanogaster. Our analyses uncover longitudinal population structure, provide evidence for continent-wide selective sweeps, identify candidate genes for local climate adaptation, and document clines in chromosomal inversion and transposable element frequencies. We also characterize variation among populations in the composition of the fly microbiome, and identify five new DNA viruses in our samples.
Keywords: SNPs; adaptation, demography; clines; population genomics; selection; structural variants.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Anderson AR, Hoffmann AA, McKechnie SW, Umina PA, Weeks AR.. 2005. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol Ecol. 14(3):851–858. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

