Expansion of GGC Repeat in GIPC1 Is Associated with Oculopharyngodistal Myopathy
- PMID: 32413282
- PMCID: PMC7273532
- DOI: 10.1016/j.ajhg.2020.04.011
Expansion of GGC Repeat in GIPC1 Is Associated with Oculopharyngodistal Myopathy
Abstract
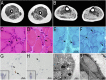
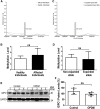
Oculopharyngodistal myopathy (OPDM) is an adult-onset inherited neuromuscular disorder characterized by progressive ptosis, external ophthalmoplegia, and weakness of the masseter, facial, pharyngeal, and distal limb muscles. The myopathological features are presence of rimmed vacuoles (RVs) in the muscle fibers and myopathic changes of differing severity. Inheritance is variable, with either putative autosomal-dominant or autosomal-recessive pattern. Here, using a comprehensive strategy combining whole-genome sequencing (WGS), long-read whole-genome sequencing (LRS), linkage analysis, repeat-primed polymerase chain reaction (RP-PCR), and fluorescence amplicon length analysis polymerase chain reaction (AL-PCR), we identified an abnormal GGC repeat expansion in the 5' UTR of GIPC1 in one out of four families and three sporadic case subjects from a Chinese OPDM cohort. Expanded GGC repeats were further confirmed as the cause of OPDM in an additional 2 out of 4 families and 6 out of 13 sporadic Chinese individuals with OPDM, as well as 7 out of 194 unrelated Japanese individuals with OPDM. Methylation, qRT-PCR, and western blot analysis indicated that GIPC1 mRNA levels were increased while protein levels were unaltered in OPDM-affected individuals. RNA sequencing indicated p53 signaling, vascular smooth muscle contraction, ubiquitin-mediated proteolysis, and ribosome pathways were involved in the pathogenic mechanisms of OPDM-affected individuals with GGC repeat expansion in GIPC1. This study provides further evidence that OPDM is associated with GGC repeat expansions in distinct genes and highly suggests that expanded GGC repeat units are essential in the pathogenesis of OPDM, regardless of the genes in which the expanded repeats are located.
Keywords: GGC repeat expansions; GIPC1; RNA-seq; intranuclear inclusions; oculopharyngodistal myopathy.
Copyright © 2020 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Durmus H., Laval S.H., Deymeer F., Parman Y., Kiyan E., Gokyigiti M., Ertekin C., Ercan I., Solakoglu S., Karcagi V. Oculopharyngodistal myopathy is a distinct entity: clinical and genetic features of 47 patients. Neurology. 2011;76:227–235. - PubMed
-
- Satoyoshi E., Kinoshita M. Oculopharyngodistal myopathy. Arch. Neurol. 1977;34:89–92. - PubMed
-
- Lu H., Luan X., Yuan Y., Dong M., Sun W., Yan C. The clinical and myopathological features of oculopharyngodistal myopathy in a Chinese family. Neuropathology. 2008;28:599–603. - PubMed
-
- Minami N., Ikezoe K., Kuroda H., Nakabayashi H., Satoyoshi E., Nonaka I. Oculopharyngodistal myopathy is genetically heterogeneous and most cases are distinct from oculopharyngeal muscular dystrophy. Neuromuscul. Disord. 2001;11:699–702. - PubMed
-
- Mignarri A., Carluccio M.A., Malandrini A., Sicurelli F., Galli L., Mazzei M.A., Federico A., Orrico A., Dotti M.T. The first Italian patient with oculopharyngodistal myopathy: case report and considerations on differential diagnosis. Neuromuscul. Disord. 2012;22:759–762. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

