Characterizing the Causal Pathway for Genetic Variants Associated with Neurological Phenotypes Using Human Brain-Derived Proteome Data
- PMID: 32413284
- PMCID: PMC7273531
- DOI: 10.1016/j.ajhg.2020.04.007
Characterizing the Causal Pathway for Genetic Variants Associated with Neurological Phenotypes Using Human Brain-Derived Proteome Data
Abstract
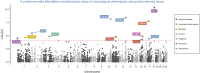
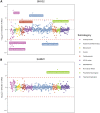
Leveraging high-dimensional molecular datasets can help us develop mechanistic insight into associations between genetic variants and complex traits. In this study, we integrated human proteome data derived from brain tissue to evaluate whether targeted proteins putatively mediate the effects of genetic variants on seven neurological phenotypes (Alzheimer disease, amyotrophic lateral sclerosis, depression, insomnia, intelligence, neuroticism, and schizophrenia). Applying the principles of Mendelian randomization (MR) systematically across the genome highlighted 43 effects between genetically predicted proteins derived from the dorsolateral prefrontal cortex and these outcomes. Furthermore, genetic colocalization provided evidence that the same causal variant at 12 of these loci was responsible for variation in both protein and neurological phenotype. This included genes such as DCC, which encodes the netrin-1 receptor and has an important role in the development of the nervous system (p = 4.29 × 10-11 with neuroticism), as well as SARM1, which has been previously implicated in axonal degeneration (p = 1.76 × 10-08 with amyotrophic lateral sclerosis). We additionally conducted a phenome-wide MR study for each of these 12 genes to assess potential pleiotropic effects on 700 complex traits and diseases. Our findings suggest that genes such as SNX32, which was initially associated with increased risk of Alzheimer disease, may potentially influence other complex traits in the opposite direction. In contrast, genes such as CTSH (which was also associated with Alzheimer disease) and SARM1 may make worthwhile therapeutic targets because they did not have genetically predicted effects on any of the other phenotypes after correcting for multiple testing.
Keywords: Mendelian randomization; brain-derived proteins; cognitive; genetic colocalization; neurological; phenome-wide association study; protein quantitative trait loci; psychaitric.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
T.R.G. and C.L.R have previously received research funding from Sanofi. T.G.R. and N.K. have both been previously funded by Sanofi. T.R.G receives research funding from GlaxoSmithKline and Biogen. However, none of these factors contributed to the design or analysis of this study.
Figures

References
-
- Trenkmann M. Lessons from 1 million genomes. Nat. Rev. Genet. 2018;19:592–593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

