Linking Virus Discovery to Immune Responses Visualized during Zebrafish Infections
- PMID: 32413307
- PMCID: PMC7854381
- DOI: 10.1016/j.cub.2020.04.031
Linking Virus Discovery to Immune Responses Visualized during Zebrafish Infections
Abstract
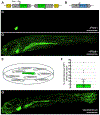
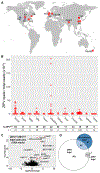
The discovery of new viruses currently outpaces our capacity for experimental examination of infection biology. To better couple virus discovery with immunology, we genetically modified zebrafish to visually report on virus infections. After generating a strain that expresses green fluorescent protein (GFP) under an interferon-stimulated gene promoter, we repeatedly observed transgenic larvae spontaneously expressing GFP days after hatching. RNA sequencing comparisons of co-housed GFP-positive and GFP-negative zebrafish revealed a naturally occurring picornavirus that induced a canonical interferon-mediated response and hundreds of antiviral defense genes not observed following immunostimulatory treatments or experimental infections with other viruses. Among the many genes induced by picornavirus infection was a large set encoding guanosine triphosphatase (GTPase) of immunity-associated proteins (GIMAPs). The GIMAP gene family is massively expanded in fish genomes and may also play a crucial role in antiviral responses in mammals, including humans. We subsequently detected zebrafish picornavirus in publicly available sequencing data from seemingly asymptomatic zebrafish in many research institutes and found that it altered gene expression in a previous study of zebrafish development. Experiments revealed a horizontal mode of virus transmission, highlighting a system for studying the spread of picornavirus infections within and between individuals. Our study describes a naturally occurring picornavirus that elicits strong antiviral responses in zebrafish and provides new strategies for simultaneously discovering viruses and their impact on vertebrate hosts.
Keywords: GIMAP; ISG15; evolution; host-pathogen; immunity; infection; interferon; picornavirus; virus; zebrafish.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




References
-
- Cobian Guemes AG, Youle M, Cantu VA, Felts B, Nulton J, and Rohwer F (2016). Viruses as Winners in the Game of Life. Annu. Rev. Virol 3, 197–214. - PubMed
-
- Suttle CA (2005). Viruses in the sea. Nature 437, 356–361. - PubMed
-
- Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X, Qin X-C, Li J, Cao J-P, Eden J-S, et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. - PubMed
-
- Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, Li K, Wang W, Eden J-S, Shen J-J, Liu L, et al. (2018). The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

