LINC00261 Is Differentially Expressed in Pancreatic Cancer Subtypes and Regulates a Pro-Epithelial Cell Identity
- PMID: 32414223
- PMCID: PMC7281485
- DOI: 10.3390/cancers12051227
LINC00261 Is Differentially Expressed in Pancreatic Cancer Subtypes and Regulates a Pro-Epithelial Cell Identity
Abstract
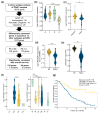
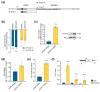
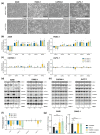
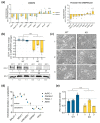
Pancreatic adenocarcinoma (PDAC) is one of the major causes of cancer-associated deaths worldwide, with a dismal prognosis that has not significantly changed over the last decades. Transcriptional analysis has provided valuable insights into pancreatic tumorigenesis. Specifically, pancreatic cancer subtypes were identified, characterized by specific mutations and gene expression changes associated with differences in patient survival. In addition to differentially regulated mRNAs, non-coding RNAs, including long non-coding RNAs (lncRNAs), were shown to have subtype-specific expression patterns. Hence, we aimed to characterize prognostic lncRNAs with deregulated expression in the squamous subtype of PDAC, which has the worst prognosis. Extensive in silico analyses followed by in vitro experiments identified long intergenic non-coding RNA 261 (LINC00261) as a downregulated lncRNA in the squamous subtype of PDAC, which is generally associated with transforming growth factor β (TGFβ) signaling in human cancer cells. Its genomic neighbor, the transcription factor forkhead box protein A2 (FOXA2), regulated LINC00261 expression by direct binding of the LINC00261 promoter. CRISPR-mediated knockdown and promoter knockout validated the importance of LINC00261 in TGFβ-mediated epithelial-mesenchymal transition (EMT) and established the epithelial marker E-cadherin, an important cell adhesion protein, as a downstream target of LINC00261. Consequently, depletion of LINC00261 enhanced motility and invasiveness of PANC-1 cells in vitro. Altogether, our data suggest that LINC00261 is an important tumor-suppressive lncRNA in PDAC that is involved in maintaining a pro-epithelial state associated with favorable disease outcome.
Keywords: CDH1; EMT; FOXA2; LINC00261; PDAC; TGFβ; lncRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

