Unique properties of a subset of human pluripotent stem cells with high capacity for self-renewal
- PMID: 32415101
- PMCID: PMC7229198
- DOI: 10.1038/s41467-020-16214-8
Unique properties of a subset of human pluripotent stem cells with high capacity for self-renewal
Abstract
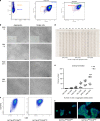
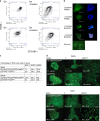
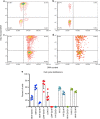
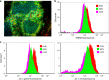
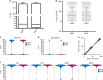
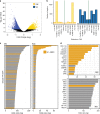
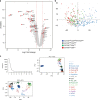
Archetypal human pluripotent stem cells (hPSC) are widely considered to be equivalent in developmental status to mouse epiblast stem cells, which correspond to pluripotent cells at a late post-implantation stage of embryogenesis. Heterogeneity within hPSC cultures complicates this interspecies comparison. Here we show that a subpopulation of archetypal hPSC enriched for high self-renewal capacity (ESR) has distinct properties relative to the bulk of the population, including a cell cycle with a very low G1 fraction and a metabolomic profile that reflects a combination of oxidative phosphorylation and glycolysis. ESR cells are pluripotent and capable of differentiation into primordial germ cell-like cells. Global DNA methylation levels in the ESR subpopulation are lower than those in mouse epiblast stem cells. Chromatin accessibility analysis revealed a unique set of open chromatin sites in ESR cells. RNA-seq at the subpopulation and single cell levels shows that, unlike mouse epiblast stem cells, the ESR subset of hPSC displays no lineage priming, and that it can be clearly distinguished from gastrulating and extraembryonic cell populations in the primate embryo. ESR hPSC correspond to an earlier stage of post-implantation development than mouse epiblast stem cells.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

