Large-scale network analysis captures biological features of bacterial plasmids
- PMID: 32415210
- PMCID: PMC7229196
- DOI: 10.1038/s41467-020-16282-w
Large-scale network analysis captures biological features of bacterial plasmids
Abstract
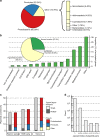
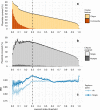
Many bacteria can exchange genetic material through horizontal gene transfer (HGT) mediated by plasmids and plasmid-borne transposable elements. Here, we study the population structure and dynamics of over 10,000 bacterial plasmids, by quantifying their genetic similarities and reconstructing a network based on their shared k-mer content. We use a community detection algorithm to assign plasmids into cliques, which correlate with plasmid gene content, bacterial host range, GC content, and existing classifications based on replicon and mobility (MOB) types. Further analysis of plasmid population structure allows us to uncover candidates for yet undescribed replicon genes, and to identify transposable elements as the main drivers of HGT at broad phylogenetic scales. Our work illustrates the potential of network-based analyses of the bacterial 'mobilome' and opens up the prospect of a natural, exhaustive classification framework for bacterial plasmids.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Shintani, M. & Suzuki, H. in DNA Traffic in the Environment 109–133 (Springer, Singapore, 2019).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

