Multiscale Coarse-Grained Approach to Investigate Self-Association of Antibodies
- PMID: 32416079
- PMCID: PMC7264848
- DOI: 10.1016/j.bpj.2020.04.022
Multiscale Coarse-Grained Approach to Investigate Self-Association of Antibodies
Abstract
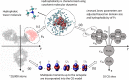
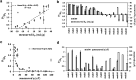
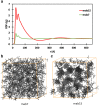
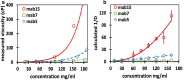
Self-association of therapeutic monoclonal antibodies (mabs) are thought to modulate the undesirably high viscosity observed in their concentrated solutions. Computational prediction of such a self-association behavior is advantageous early during mab drug candidate selection when material availability is limited. Here, we present a coarse-grained (CG) simulation method that enables microsecond molecular dynamics simulations of full-length antibodies at high concentrations. The proposed approach differs from others in two ways: first, charges are assigned to CG beads in an effort to reproduce molecular multipole moments and charge asymmetry of full-length antibodies instead of only localized charges. This leads to great improvements in the agreement between CG and all-atom electrostatic fields. Second, the distinctive hydrophobic character of each antibody is incorporated through empirical adjustments to the short-range van der Waals terms dictated by cosolvent all-atom molecular dynamics simulations of antibody variable regions. CG simulations performed on a set of 15 different mabs reveal that diffusion coefficients in crowded environments are markedly impacted by intermolecular interactions. Diffusion coefficients computed from the simulations are in correlation with experimentally measured observables, including viscosities at a high concentration. Further, we show that the evaluation of electrostatic and hydrophobic characters of the mabs is useful in predicting the nonuniform effect of salt on the viscosity of mab solutions. This CG modeling approach is particularly applicable as a material-free screening tool for selecting antibody candidates with desirable viscosity properties.
Copyright © 2020 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Carter P.J. Potent antibody therapeutics by design. Nat. Rev. Immunol. 2006;6:343–357. - PubMed
-
- Lobo E.D., Hansen R.J., Balthasar J.P. Antibody pharmacokinetics and pharmacodynamics. J. Pharm. Sci. 2004;93:2645–2668. - PubMed
-
- Yadav S., Shire S.J., Kalonia D.S. Factors affecting the viscosity in high concentration solutions of different monoclonal antibodies. J. Pharm. Sci. 2010;99:4812–4829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

