SARS-CoV-2 detection by direct rRT-PCR without RNA extraction
- PMID: 32416598
- PMCID: PMC7204723
- DOI: 10.1016/j.jcv.2020.104423
SARS-CoV-2 detection by direct rRT-PCR without RNA extraction
Abstract
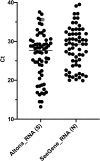
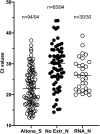
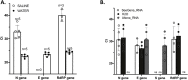
Rapid and reliable screening of SARS-CoV-2 is fundamental to assess viral spread and limit the pandemic we are facing. In this study, we compared direct rRT-PCR method (without RNA extraction) using SeeGene AllplexTM 2019-nCoV rRT-PCR with the RealStar® SARS-CoV-2 rRT-PCR kit (Altona Diagnostics). Furthermore, we assessed the impact of swab storage media composition on PCR efficiency. We show that SeeGene and Altona's assays provide similar efficiency. Importantly, we provide evidence that RNA extraction can be successfully bypassed when samples are stored in UTM medium or in molecular water but not when samples are stored in saline solution and in Hanks medium.
Keywords: COVID19; Coronavirus; Direct rRTPCR; RNA extraction; SARS-CoV-2; Virus detection.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest None.
Figures

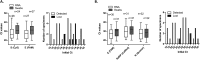
Similar articles
-
A simple method for SARS-CoV-2 detection by rRT-PCR without the use of a commercial RNA extraction kit.J Virol Methods. 2020 Nov;285:113960. doi: 10.1016/j.jviromet.2020.113960. Epub 2020 Aug 22. J Virol Methods. 2020. PMID: 32835738 Free PMC article.
-
Value of swab types and collection time on SARS-COV-2 detection using RT-PCR assay.J Virol Methods. 2020 Dec;286:113974. doi: 10.1016/j.jviromet.2020.113974. Epub 2020 Sep 16. J Virol Methods. 2020. PMID: 32949663 Free PMC article.
-
Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits.J Clin Virol. 2020 Aug;129:104519. doi: 10.1016/j.jcv.2020.104519. Epub 2020 Jun 23. J Clin Virol. 2020. PMID: 32629187 Free PMC article.
-
Laboratory diagnosis of SARS-CoV-2 - A review of current methods.J Infect Public Health. 2020 Jul;13(7):901-905. doi: 10.1016/j.jiph.2020.06.005. Epub 2020 Jun 7. J Infect Public Health. 2020. PMID: 32534946 Free PMC article. Review.
-
SARS-CoV-2 detection in different respiratory sites: A systematic review and meta-analysis.EBioMedicine. 2020 Sep;59:102903. doi: 10.1016/j.ebiom.2020.102903. Epub 2020 Jul 24. EBioMedicine. 2020. PMID: 32718896 Free PMC article.
Cited by
-
Direct Lysis RT-qPCR of SARS-CoV-2 in Cell Culture Supernatant Allows for Fast and Accurate Quantification.Viruses. 2022 Feb 28;14(3):508. doi: 10.3390/v14030508. Viruses. 2022. PMID: 35336915 Free PMC article.
-
COVID-19 laboratory diagnosis: comparative analysis of different RNA extraction methods for SARS-CoV-2 detection by two amplification protocols.Rev Inst Med Trop Sao Paulo. 2021 Jun 25;63:e52. doi: 10.1590/S1678-9946202163052. eCollection 2021. Rev Inst Med Trop Sao Paulo. 2021. PMID: 34190954 Free PMC article.
-
A simple method for SARS-CoV-2 detection by rRT-PCR without the use of a commercial RNA extraction kit.J Virol Methods. 2020 Nov;285:113960. doi: 10.1016/j.jviromet.2020.113960. Epub 2020 Aug 22. J Virol Methods. 2020. PMID: 32835738 Free PMC article.
-
Comparative Performance of a New SARS-CoV-2 Rapid Detection System.Microbiol Spectr. 2021 Oct 31;9(2):e0020521. doi: 10.1128/Spectrum.00205-21. Epub 2021 Oct 13. Microbiol Spectr. 2021. PMID: 34643409 Free PMC article.
-
High-Throughput COVID-19 Testing of Naso-Oropharyngeal Swabs Using a Sensitive Extraction-Free Sample Preparation Method.Microbiol Spectr. 2022 Aug 31;10(4):e0135822. doi: 10.1128/spectrum.01358-22. Epub 2022 Aug 11. Microbiol Spectr. 2022. PMID: 35950846 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

