ICTV Virus Taxonomy Profile: Hepadnaviridae
- PMID: 32416744
- PMCID: PMC7414443
- DOI: 10.1099/jgv.0.001415
ICTV Virus Taxonomy Profile: Hepadnaviridae
Abstract
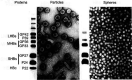
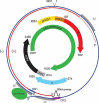
The family Hepadnaviridae comprises small enveloped viruses with a partially double-stranded DNA genome of 3.0-3.4 kb. All family members express three sets of proteins (preC/C, polymerase and preS/S) and replication involves reverse transcription within nucleocapsids in the cytoplasm of hepatocytes. Hepadnaviruses are hepatotropic and infections may be transient or persistent. There are five genera: Parahepadnavirus, Metahepadnavirus, Herpetohepadnavirus, Avihepadnavirus and Orthohepadnavirus. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Hepadnaviridae, which is available at ictv.global/report/hepadnaviridae.
Keywords: Hepadnaviridae; ICTV Report; hepatitis B virus; taxonomy.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

