Biochemical characterization of SARS-CoV-2 nucleocapsid protein
- PMID: 32416961
- PMCID: PMC7190499
- DOI: 10.1016/j.bbrc.2020.04.136
Biochemical characterization of SARS-CoV-2 nucleocapsid protein
Erratum in
-
Corrigendum to "Biochemical characterization of SARS-CoV-2 nucleocapsid protein".Biochem Biophys Res Commun. 2022 Jul 23;614:225. doi: 10.1016/j.bbrc.2022.05.058. Epub 2022 May 25. Biochem Biophys Res Commun. 2022. PMID: 35643631 Free PMC article. No abstract available.
Abstract
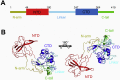
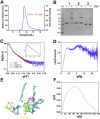
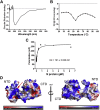
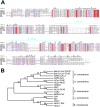
The nucleocapsid (N) protein is an important antigen for coronavirus, which participate in RNA package and virus particle release. In this study, we expressed the N protein of SARS-CoV-2 and characterized its biochemical properties. Static light scattering, size exclusive chromatography, and small-angle X-ray scattering (SAXS) showed that the purified N protein is largely a dimer in solution. CD spectra showed that it has a high percentage of disordered region at room temperature while it was best structured at 55 °C, suggesting its structural dynamics. Fluorescence polarization assay showed it has non-specific nucleic acid binding capability, which raised a concern in using it as a diagnostic marker. Immunoblot assays confirmed the presence of IgA, IgM and IgG antibodies against N antigen in COVID-19 infection patients' sera, proving the importance of this antigen in host immunity and diagnostics.
Keywords: Antigenicity; COVID-19; Nucleocapsid protein; SARS-CoV-2; SAXS; Structure and function.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- WHO Novel coronavirus – China. http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ accessed Jan 19, 2020.
-
- WHO Coronavirus (COVID-19) https://covid19.who.int/
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England) 2020;395:565–574. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

