Circles of Life: linking metabolic and epigenetic cycles to immunity
- PMID: 32418209
- PMCID: PMC7576883
- DOI: 10.1111/imm.13207
Circles of Life: linking metabolic and epigenetic cycles to immunity
Abstract
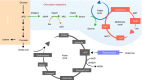
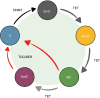
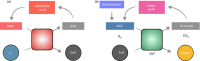
Metabolites are the essential substrates for epigenetic modification enzymes to write or erase the epigenetic blueprint in cells. Hence, the availability of nutrients and activity of metabolic pathways strongly influence the enzymatic function. Recent studies have shed light on the choreography between metabolome and epigenome in the control of immune cell differentiation and function, with a major focus on histone modifications. Yet, despite its importance in gene regulation, DNA methylation and its relationship with metabolism is relatively unclear. In this review, we will describe how the metabolic flux can influence epigenetic networks in innate and adaptive immune cells, with a focus on the DNA methylation cycle and the metabolites S-adenosylmethionine and α-ketoglutarate. Future directions will be discussed for this rapidly emerging field.
Keywords: 5-hydroxymethylcytosine; B cells; DNA methylation; DNA methyltransferases; Krebs cycle; T cells; epigenetics; immunometabolism; macrophages; mitochondria; one-carbon metabolism; ten-eleven translocation.
© 2020 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
References
-
- Li X, Wenes M, Romero P, Huang SC‐C, Fendt S‐M, Ho P‐C. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat Rev Clin Oncol (R Coll Radiol) 2019; 16:425–41. - PubMed
-
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007; 128:635–8. - PubMed
-
- Álvarez‐Errico D, Vento‐Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol 2015; 15:7–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CLIP/CRI/Cancer Research Institute/United States
- Independent Investigator Fund (La Jolla Institute/Kyowa Kirin)
- IRG-16-186-21/Case Comprehensive Cancer Center American Cancer Society Pilot Grants
- IRG-91-022-19/Case Comprehensive Cancer Center American Cancer Society Pilot Grants
- K22 CA241290/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources

