Comprehensive analysis of competing endogenous RNA network in Wilms tumor based on the TARGET database
- PMID: 32420153
- PMCID: PMC7214997
- DOI: 10.21037/tau.2020.01.34
Comprehensive analysis of competing endogenous RNA network in Wilms tumor based on the TARGET database
Abstract
Background: Wilms tumor (WT) was the most common malignant tumor of urinary system in children. With the advances in gene sequencing, research of molecular mechanism of WT tumor was gradually increasing. However, few studies have explored the influence of competing endogenous RNA (ceRNA) in WT. Accordingly, we aimed to explore the mechanisms of ceRNA co-expression network in WT.
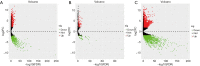
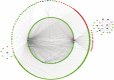
Methods: A total of 6 non-tumor controls and 127 WT patients' RNA-seq data combined with clinical data was acquired from Therapeutically Applicable Research to Generate Effective Treatments (TARGET) database. Differentially expressed lncRNA, miRNA and mRNA between WT tissues and normal tissues were analyzed using "edgeR" package in R software. Weighted gene co-expression network analysis (WGCNA) was utilized to construct the ceRNA co-expression network while Molecular Complex Detection (MCODE) algorithm was used to extract the pivotal sub-network. Function annotation of mRNA was performed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Survival analysis was then conducted based on long-rank test and Kaplan-Meier curves using the survival package.
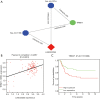
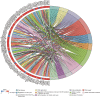
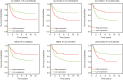
Results: By applying the "edgeR" package in R, the transcriptome expression data of 127 WT tissues with 6 normal tissues were normalized. Moreover, 146 DElncRNAs, 62 DEmiRNAs, 287 DEmRNAs of them were involved in ceRNA network after applying WGCNA. According to MCODE, we identified that the interactions between LINC002253 (lncRNA) and TRIM71 (mRNA) was mediated by hsa-mir-301a and hsa-mir-301b (miRNA). Furthermore, we detected 13 DElncRNAs which were significantly associated with the progression of WT.
Conclusions: We used WGCNA method to construct the WT ceRNA network for the first time. TRIM71 was identified to be the most closely related genes involved in hub sub-network by MCDOE, suggesting it might act as a new drug target and prognostic factor based on our comprehensive results.
Keywords: Therapeutically Applicable Research to Generate Effective Treatments (TARGET); Wilms tumor (WT); competitive endogenous RNA (ceRNA); lncRNA.
2020 Translational Andrology and Urology. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.01.34). The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Comprehensive Analysis of a Long Noncoding RNA-Associated Competing Endogenous RNA Network in Wilms Tumor.Cancer Control. 2020 Apr-Jun;27(2):1073274820936991. doi: 10.1177/1073274820936991. Cancer Control. 2020. PMID: 32597194 Free PMC article.
-
Comprehensive Analysis of lncRNA-Mediated ceRNA Crosstalk and Identification of Prognostic Biomarkers in Wilms' Tumor.Biomed Res Int. 2020 Feb 21;2020:4951692. doi: 10.1155/2020/4951692. eCollection 2020. Biomed Res Int. 2020. PMID: 32149111 Free PMC article.
-
[Construction and analysis of competitive endogenous RNA regulatory network related to gastric cancer].Zhonghua Zhong Liu Za Zhi. 2020 Feb 23;42(2):115-121. doi: 10.3760/cma.j.issn.0253-3766.2020.02.006. Zhonghua Zhong Liu Za Zhi. 2020. PMID: 32135645 Chinese.
-
Identification of Potential Biomarkers of Prognosis-Related Long Non-Coding RNA (lncRNA) in Pediatric Rhabdoid Tumor of the Kidney Based on ceRNA Networks.Med Sci Monit. 2020 Dec 17;26:e927725. doi: 10.12659/MSM.927725. Med Sci Monit. 2020. PMID: 33328429 Free PMC article.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
References
LinkOut - more resources
Full Text Sources
