Distinguishing Kawasaki Disease from Febrile Infectious Disease Using Gene Pair Signatures
- PMID: 32420360
- PMCID: PMC7201505
- DOI: 10.1155/2020/6539398
Distinguishing Kawasaki Disease from Febrile Infectious Disease Using Gene Pair Signatures
Abstract
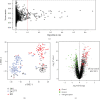
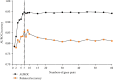
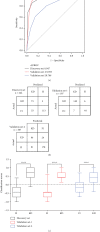
Kawasaki disease (KD) is an acute systemic vasculitis of childhood with prolonged fever, and the diagnosis of KD is mainly based on clinical criteria, which is prone to misdiagnosis with other febrile infectious (FI) diseases. Currently, there remain no effective molecular markers for KD diagnosis. In this study, we aimed to use a relative-expression-based method k-TSP and resampling framework to identify robust gene pair signatures to distinguish KD from bacterial and virus febrile infectious diseases. Our study pool consisted of 808 childhood patients from several studies and assigned to three groups, namely, the discovery set (n = 224), validation set-1 (n = 197), and validation set-2 (n = 387). We had identified 60 biologically relevant gene pairs and developed a top-ranked gene pair classifier (TRGP) using the first seven signatures, with the area under the receiver-operating characteristic curves (AUROC) of 0.947 (95% CI, 0.918-0.976), a sensitivity of 0.936 (95% CI, 0.872-0.987), and a specificity of 0.774 (95% CI, 0.705-0.836) in the discovery set. In the validation set-1, the TRGP classifier distinguished KD from FI with AUROC of 0.955 (95% CI, 0.919-0.991), a sensitivity of 0.959 (95% CI, 0.925-0.986), and a specificity of 0.863 (95% CI, 0.764-0.961). In the validation set-2, the predictive performance of classification was with an AUROC of 0.796 (95% CI, 0.747-0.845), a sensitivity of 0.797 (95% CI, 0.720-0.864), and a specificity of 0.661 (95% CI, 0.606-0.717). Our study reveals that gene pair signatures are robust across diverse studies and can be utilized as objective biomarkers to distinguish KD from FI, helping to develop a fast, simple, and effective molecular approach to improve the diagnosis of KD.
Copyright © 2020 Jiayong Zhong et al.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Diagnosis of Kawasaki Disease Using a Minimal Whole-Blood Gene Expression Signature.JAMA Pediatr. 2018 Oct 1;172(10):e182293. doi: 10.1001/jamapediatrics.2018.2293. Epub 2018 Oct 1. JAMA Pediatr. 2018. PMID: 30083721 Free PMC article.
-
Circulating microRNAs differentiate Kawasaki Disease from infectious febrile illnesses in childhood.J Mol Cell Cardiol. 2020 Sep;146:12-18. doi: 10.1016/j.yjmcc.2020.06.011. Epub 2020 Jul 4. J Mol Cell Cardiol. 2020. PMID: 32634388
-
Diagnostic Test Accuracy of a 2-Transcript Host RNA Signature for Discriminating Bacterial vs Viral Infection in Febrile Children.JAMA. 2016 Aug 23-30;316(8):835-45. doi: 10.1001/jama.2016.11236. JAMA. 2016. PMID: 27552617 Free PMC article.
-
Identification of susceptibility genes for Kawasaki disease.Nihon Rinsho Meneki Gakkai Kaishi. 2010;33(2):73-80. doi: 10.2177/jsci.33.73. Nihon Rinsho Meneki Gakkai Kaishi. 2010. PMID: 20453442 Review.
-
Promising biomarkers of Kawasaki disease: markers that aid in diagnosis.Expert Rev Mol Diagn. 2024 Nov 24:1-13. doi: 10.1080/14737159.2024.2432025. Online ahead of print. Expert Rev Mol Diagn. 2024. PMID: 39556196 Review.
Cited by
-
Host Blood Gene Signatures Can Detect the Progression to Severe and Cerebral Malaria.Front Cell Infect Microbiol. 2021 Oct 22;11:743616. doi: 10.3389/fcimb.2021.743616. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34746025 Free PMC article.
-
Identifying the risk of Kawasaki disease based solely on routine blood test features through novel construction of machine learning models.Comput Struct Biotechnol J. 2025 Jun 25;27:2832-2842. doi: 10.1016/j.csbj.2025.06.037. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40677244 Free PMC article.
-
Applications of gene pair methods in clinical research: advancing precision medicine.Mol Biomed. 2025 Apr 9;6(1):22. doi: 10.1186/s43556-025-00263-w. Mol Biomed. 2025. PMID: 40202606 Free PMC article. Review.
References
-
- Kawasaki T. Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children. Arerugī. 1967;16:178–222. - PubMed
-
- McCrindle B. W., Rowley A. H., Newburger J. W., et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation. 2017;135(17):e927–e999. doi: 10.1161/cir.0000000000000484. On behalf of the American Heart Association Rheumatic Fever, Endocarditis, and Kawasaki Disease Committee of the Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; Council on Cardiovascular Surgery and Anesthesia; and Council on Epidemiology and Prevention. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

