Sequence-structure-function relationships in class I MHC: A local frustration perspective
- PMID: 32421728
- PMCID: PMC7233585
- DOI: 10.1371/journal.pone.0232849
Sequence-structure-function relationships in class I MHC: A local frustration perspective
Abstract
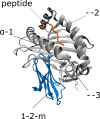
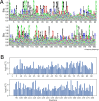
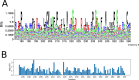
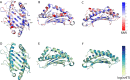
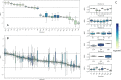
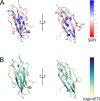
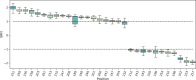
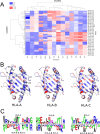
Class I Major Histocompatibility Complex (MHC) binds short antigenic peptides with the help of Peptide Loading Complex (PLC), and presents them to T-cell Receptors (TCRs) of cytotoxic T-cells and Killer-cell Immunglobulin-like Receptors (KIRs) of Natural Killer (NK) cells. With more than 10000 alleles, human MHC (Human Leukocyte Antigen, HLA) is the most polymorphic protein in humans. This allelic diversity provides a wide coverage of peptide sequence space, yet does not affect the three-dimensional structure of the complex. Moreover, TCRs mostly interact with HLA in a common diagonal binding mode, and KIR-HLA interaction is allele-dependent. With the aim of establishing a framework for understanding the relationships between polymorphism (sequence), structure (conserved fold) and function (protein interactions) of the human MHC, we performed here a local frustration analysis on pMHC homology models covering 1436 HLA I alleles. An analysis of local frustration profiles indicated that (1) variations in MHC fold are unlikely due to minimally-frustrated and relatively conserved residues within the HLA peptide-binding groove, (2) high frustration patches on HLA helices are either involved in or near interaction sites of MHC with the TCR, KIR, or tapasin of the PLC, and (3) peptide ligands mainly stabilize the F-pocket of HLA binding groove.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

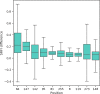
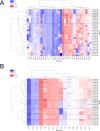
References
-
- Bahar I, Jernigan RL, Dill KA. Protein actions: principles and modeling. 2017. Available: https://www.crcpress.com/Protein-Actions-Principles-and-Modeling/Bahar-J...
-
- Kimura M, Ohta T. On some principles governing molecular evolution. Proc Natl Acad Sci U S A. 1974;71: 2848–52. Available: http://www.ncbi.nlm.nih.gov/pubmed/4527913 10.1073/pnas.71.7.2848 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

