Che-1/AATF binds to RNA polymerase I machinery and sustains ribosomal RNA gene transcription
- PMID: 32421830
- PMCID: PMC7293028
- DOI: 10.1093/nar/gkaa344
Che-1/AATF binds to RNA polymerase I machinery and sustains ribosomal RNA gene transcription
Abstract
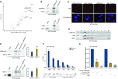
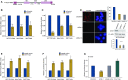
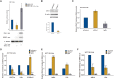
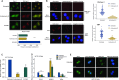
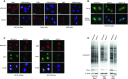
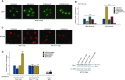
Originally identified as an RNA polymerase II interactor, Che-1/AATF (Che-1) has now been recognized as a multifunctional protein involved in cell-cycle regulation and cancer progression, as well as apoptosis inhibition and response to stress. This protein displays a peculiar nucleolar localization and it has recently been implicated in pre-rRNA processing and ribosome biogenesis. Here, we report the identification of a novel function of Che-1 in the regulation of ribosomal RNA (rRNA) synthesis, in both cancer and normal cells. We demonstrate that Che-1 interacts with RNA polymerase I and nucleolar upstream binding factor (UBF) and promotes RNA polymerase I-dependent transcription. Furthermore, this protein binds to the rRNA gene (rDNA) promoter and modulates its epigenetic state by contrasting the recruitment of HDAC1. Che-1 downregulation affects RNA polymerase I and UBF recruitment on rDNA and leads to reducing rDNA promoter activity and 47S pre-rRNA production. Interestingly, Che-1 depletion induces abnormal nucleolar morphology associated with re-distribution of nucleolar proteins. Finally, we show that upon DNA damage Che-1 re-localizes from rDNA to TP53 gene promoter to induce cell-cycle arrest. This previously uncharacterized function of Che-1 confirms the important role of this protein in the regulation of ribosome biogenesis, cellular proliferation and response to stress.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

