Fam20C regulates protein secretion by Cab45 phosphorylation
- PMID: 32422653
- PMCID: PMC7265331
- DOI: 10.1083/jcb.201910089
Fam20C regulates protein secretion by Cab45 phosphorylation
Abstract
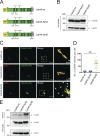
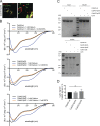
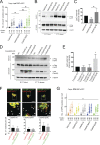
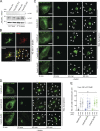
The TGN is a key compartment for the sorting and secretion of newly synthesized proteins. At the TGN, soluble proteins are sorted based on the instructions carried in their oligosaccharide backbones or by a Ca2+-mediated process that involves the cargo-sorting protein Cab45. Here, we show that Cab45 is phosphorylated by the Golgi-specific protein kinase Fam20C. Mimicking of phosphorylation translocates Cab45 into TGN-derived vesicles, which goes along with an increased export of LyzC, a Cab45 client. Our findings demonstrate that Fam20C plays a key role in the export of Cab45 clients by fine-tuning Cab45 oligomerization and thus impacts Cab45 retention in the TGN.
© 2020 Hecht et al.
Figures








References
-
- Barenholz Y., and Thompson T.E.. 1980. Sphingomyelins in bilayers and biological membranes. Biochimica Et Biophysica Acta Bba - Rev Biomembr. 604:129–158. - PubMed
-
- Boncompain G., and Perez F.. 2012. Synchronising protein transport in the secretory pathway. Curr Protoc Cell Biol. 15.19.1-15.19.16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

