Connecting RNA-Modifying Similarities of TDP-43, FUS, and SOD1 with MicroRNA Dysregulation Amidst A Renewed Network Perspective of Amyotrophic Lateral Sclerosis Proteinopathy
- PMID: 32422969
- PMCID: PMC7278980
- DOI: 10.3390/ijms21103464
Connecting RNA-Modifying Similarities of TDP-43, FUS, and SOD1 with MicroRNA Dysregulation Amidst A Renewed Network Perspective of Amyotrophic Lateral Sclerosis Proteinopathy
Abstract
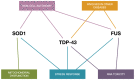
Beyond traditional approaches in understanding amyotrophic lateral sclerosis (ALS), multiple recent studies in RNA-binding proteins (RBPs)-including transactive response DNA-binding protein (TDP-43) and fused in sarcoma (FUS)-have instigated an interest in their function and prion-like properties. Given their prominence as hallmarks of a highly heterogeneous disease, this prompts a re-examination of the specific functional interrelationships between these proteins, especially as pathological SOD1-a non-RBP commonly associated with familial ALS (fALS)-exhibits similar properties to these RBPs including potential RNA-regulatory capabilities. Moreover, the cytoplasmic mislocalization, aggregation, and co-aggregation of TDP-43, FUS, and SOD1 can be identified as proteinopathies akin to other neurodegenerative diseases (NDs), eliciting strong ties to disrupted RNA splicing, transport, and stability. In recent years, microRNAs (miRNAs) have also been increasingly implicated in the disease, and are of greater significance as they are the master regulators of RNA metabolism in disease pathology. However, little is known about the role of these proteins and how they are regulated by miRNA, which would provide mechanistic insights into ALS pathogenesis. This review seeks to discuss current developments across TDP-43, FUS, and SOD1 to build a detailed snapshot of the network pathophysiology underlying ALS while aiming to highlight possible novel therapeutic targets to guide future research.
Keywords: FUS; RNA metabolism; RNA-binding proteins; SOD1; TDP-43; amyotrophic lateral sclerosis; microRNA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
"STRESSED OUT": The role of FUS and TDP-43 in amyotrophic lateral sclerosis.Int J Biochem Cell Biol. 2020 Sep;126:105821. doi: 10.1016/j.biocel.2020.105821. Epub 2020 Aug 3. Int J Biochem Cell Biol. 2020. PMID: 32758633 Review.
-
Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis.Brain Res. 2016 Sep 15;1647:65-78. doi: 10.1016/j.brainres.2016.03.036. Epub 2016 Mar 28. Brain Res. 2016. PMID: 27033831 Free PMC article. Review.
-
Rab1-dependent ER-Golgi transport dysfunction is a common pathogenic mechanism in SOD1, TDP-43 and FUS-associated ALS.Acta Neuropathol. 2015 Nov;130(5):679-97. doi: 10.1007/s00401-015-1468-2. Epub 2015 Aug 23. Acta Neuropathol. 2015. PMID: 26298469
-
Genetics of amyotrophic lateral sclerosis: A review.J Neurol Sci. 2019 Apr 15;399:217-226. doi: 10.1016/j.jns.2019.02.030. Epub 2019 Feb 21. J Neurol Sci. 2019. PMID: 30870681 Review.
-
Oxr1 improves pathogenic cellular features of ALS-associated FUS and TDP-43 mutations.Hum Mol Genet. 2015 Jun 15;24(12):3529-44. doi: 10.1093/hmg/ddv104. Epub 2015 Mar 19. Hum Mol Genet. 2015. PMID: 25792726 Free PMC article.
Cited by
-
Insights into the identification of a molecular signature for amyotrophic lateral sclerosis exploiting integrated microRNA profiling of iPSC-derived motor neurons and exosomes.Cell Mol Life Sci. 2022 Mar 14;79(3):189. doi: 10.1007/s00018-022-04217-1. Cell Mol Life Sci. 2022. PMID: 35286466 Free PMC article.
-
Induced Pluripotent Stem Cells and Their Applications in Amyotrophic Lateral Sclerosis.Cells. 2023 Mar 22;12(6):971. doi: 10.3390/cells12060971. Cells. 2023. PMID: 36980310 Free PMC article. Review.
-
Comprehensive Research on Past and Future Therapeutic Strategies Devoted to Treatment of Amyotrophic Lateral Sclerosis.Int J Mol Sci. 2022 Feb 22;23(5):2400. doi: 10.3390/ijms23052400. Int J Mol Sci. 2022. PMID: 35269543 Free PMC article. Review.
-
Mid-Gestation lethality of Atxn2l-Ablated Mice.Int J Mol Sci. 2020 Jul 20;21(14):5124. doi: 10.3390/ijms21145124. Int J Mol Sci. 2020. PMID: 32698485 Free PMC article.
-
Advancements in genetic research and RNA therapy strategies for amyotrophic lateral sclerosis (ALS): current progress and future prospects.J Neurol. 2025 Feb 26;272(3):233. doi: 10.1007/s00415-025-12975-8. J Neurol. 2025. PMID: 40009238 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

