Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture
- PMID: 32423437
- PMCID: PMC7236360
- DOI: 10.1186/s12866-020-01801-y
Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture
Abstract
Background: The microbiome of the oral cavity is the second-largest and diverse microbiota after the gut, harboring over 700 species of bacteria and including also fungi, viruses, and protozoa. With its diverse niches, the oral cavity is a very complex environment, where different microbes preferentially colonize different habitats. Recent data indicate that the oral microbiome has essential functions in maintaining oral and systemic health, and the emergence of 16S rRNA gene next-generation sequencing (NGS) has greatly contributed to revealing the complexity of its bacterial component. However, a detailed site-specific map of oral microorganisms (including also eukaryotes and viruses) and their relative abundance is still missing. Here, we aimed to obtain a comprehensive view of the healthy oral microbiome (HOM), including its drug-resistance features.
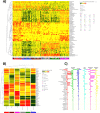
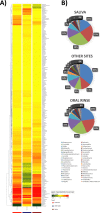
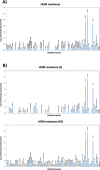
Results: The oral microbiome of twenty healthy subjects was analyzed by whole-genome sequencing (WGS) and real-time quantitative PCR microarray. Sampled oral micro-habitat included tongue dorsum, hard palate, buccal mucosa, keratinized gingiva, supragingival and subgingival plaque, and saliva with or without rinsing. Each sampled oral niche evidenced a different microbial community, including bacteria, fungi, and viruses. Alpha-diversity evidenced significant differences among the different sampled sites (p < 0.0001) but not among the enrolled subjects (p = 0.876), strengthening the notion of a recognizable HOM. Of note, oral rinse microbiome was more representative of the whole site-specific microbiomes, compared with that of saliva. Interestingly, HOM resistome included highly prevalent genes conferring resistance to macrolide, lincosamides, streptogramin, and tetracycline.
Conclusions: The data obtained in 20 subjects by WGS and microarray analysis provide for the first time a comprehensive view of HOM and its resistome, contributing to a deeper understanding of the composition of oral microbiome in the healthy subject, and providing an important reference for future studies, allowing to identify microbial signatures related to functional and metabolic alterations associated with diseases, potentially useful for targeted therapies and precision medicine.
Keywords: Oral microbiome; Resistome; Site-specific microbiome map; Whole-genome sequencing (WGS).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V, Finlay BB. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12(5):611–622. - PubMed
-
- Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137–143. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

