Molecular mechanism of the recognition of bacterially cleaved immunoglobulin by the immune regulatory receptor LILRA2
- PMID: 32424043
- PMCID: PMC7363141
- DOI: 10.1074/jbc.RA120.013354
Molecular mechanism of the recognition of bacterially cleaved immunoglobulin by the immune regulatory receptor LILRA2
Abstract
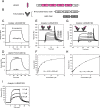
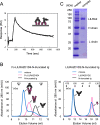
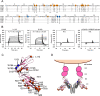
Human leukocyte immunoglobulin-like receptors (LILRs) typically regulate immune activation by binding to the human leukocyte antigen class I molecules. LILRA2, a member of the LILR family, was recently reported to bind to other unique ligands, the bacterially degraded Igs (N-truncated Igs), for the activation of immune cells. Therefore, LILRA2 is currently attracting significant attention as a novel innate immune receptor. However, the detailed recognition mechanisms required for this interaction remain unclear. In this study, using several biophysical techniques, we uncovered the molecular mechanism of N-truncated Ig recognition by LILRA2. Surface plasmon resonance analysis disclosed that LILRA2 specifically binds to N-truncated Ig with weak affinity (Kd = 4.8 μm) and fast kinetics. However, immobilized LILRA2 exhibited a significantly enhanced interaction with N-truncated Ig due to avidity effects. This suggests that cell surface-bound LILRA2 rapidly monitors and identifies bi- or multivalent abnormal N-truncated Igs through specific cross-linking to induce immune activation. Van't Hoff analysis revealed that this interaction is enthalpy-driven, with a small entropy loss, and results from differential scanning calorimetry indicated the instability of the putative LILRA2-binding site, the Fab region of the N-truncated Ig. Atomic force microscopy revealed that N truncation does not cause significant structural changes in Ig. Furthermore, mutagenesis analysis identified the hydrophobic region of LILRA2 domain 2 as the N-truncated Ig-binding site, representing a novel ligand-binding site for the LILR family. These results provide detailed insights into the molecular regulation of LILR-mediated immune responses targeting ligands that have been modified by bacteria.
Keywords: LILRA2; atomic force microscopy (AFM); bacterially cleaved immunoglobulin; cell surface receptor; immune regulation; immune regulatory receptor; immunoglobulin G (IgG); immunology; leukocyte immunoglobulin-like receptor (LILR); protein-protein interaction; surface plasmon resonance (SPR).
© 2020 Yamazaki et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Shiroishi M., Tsumoto K., Amano K., Shirakihara Y., Colonna M., Braud V. M., Allan D. S., Makadzange A., Rowland-Jones S., Willcox B., Jones E. Y., van der Merwe P. A., Kumagai I., and Maenaka K. (2003) Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. U S A 100, 8856–8861 10.1073/pnas.1431057100 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

