Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium
- PMID: 32424351
- PMCID: PMC7116672
- DOI: 10.1038/s41588-020-0624-3
Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium
Abstract
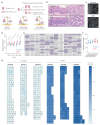
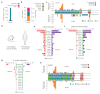
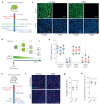
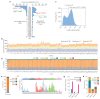
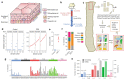
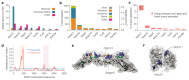
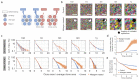
During aging, progenitor cells acquire mutations, which may generate clones that colonize the surrounding tissue. By middle age, normal human tissues, including the esophageal epithelium (EE), become a patchwork of mutant clones. Despite their relevance for understanding aging and cancer, the processes that underpin mutational selection in normal tissues remain poorly understood. Here, we investigated this issue in the esophageal epithelium of mutagen-treated mice. Deep sequencing identified numerous mutant clones with multiple genes under positive selection, including Notch1, Notch2 and Trp53, which are also selected in human esophageal epithelium. Transgenic lineage tracing revealed strong clonal competition that evolved over time. Clone dynamics were consistent with a simple model in which the proliferative advantage conferred by positively selected mutations depends on the nature of the neighboring cells. When clones with similar competitive fitness collide, mutant cell fate reverts towards homeostasis, a constraint that explains how selection operates in normal-appearing epithelium.
Conflict of interest statement
The authors declare no competing interests.
Figures








Comment in
-
Clonal competition in a confined space.Nat Genet. 2020 Jun;52(6):553-554. doi: 10.1038/s41588-020-0638-x. Nat Genet. 2020. PMID: 32424354 No abstract available.
-
Clone conflict.Nat Rev Cancer. 2020 Jul;20(7):362. doi: 10.1038/s41568-020-0278-6. Nat Rev Cancer. 2020. PMID: 32488199 No abstract available.
-
Clone wars: cells competing to win.Nat Rev Genet. 2020 Aug;21(8):446-447. doi: 10.1038/s41576-020-0256-6. Nat Rev Genet. 2020. PMID: 32504077 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_17230/MRC_/Medical Research Council/United Kingdom
- 21777/CRUK_/Cancer Research UK/United Kingdom
- 17257/CRUK_/Cancer Research UK/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
- 296194/WT_/Wellcome Trust/United Kingdom
- MC_PC_12009/MRC_/Medical Research Council/United Kingdom
- MR/S000216/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12022/9/MRC_/Medical Research Council/United Kingdom
- A21777/CRUK_/Cancer Research UK/United Kingdom
- A27326/CRUK_/Cancer Research UK/United Kingdom
- C609/A27326/CRUK_/Cancer Research UK/United Kingdom
- C57387/A21777/CRUK_/Cancer Research UK/United Kingdom
- C609/A17257/CRUK_/Cancer Research UK/United Kingdom
- A17257/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

