Inhibition of Cavin3 Degradation by the Human Parainfluenza Virus Type 2 V Protein Is Important for Efficient Viral Growth
- PMID: 32425917
- PMCID: PMC7203785
- DOI: 10.3389/fmicb.2020.00803
Inhibition of Cavin3 Degradation by the Human Parainfluenza Virus Type 2 V Protein Is Important for Efficient Viral Growth
Abstract
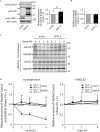
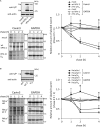
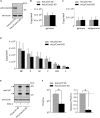
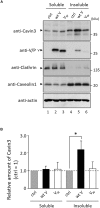
Cavin proteins have important roles in the formation of caveolae in lipid raft microdomains. Pulse-chase experiments of cells infected with human parainfluenza virus type 2 (hPIV-2) showed decreased proteasomal degradation of Cavin3. Overexpression of hPIV-2 V protein alone was sufficient to inhibit Cavin3 degradation. Immunoprecipitation analysis revealed that V protein bound to Cavin3. Trp residues within C-terminal region of V protein, as well as the N-terminal region of Cavin3, are important for V-Cavin3 interaction. Cavin3 knockdown suppressed hPIV-2 growth without affecting its entry, replication, transcription, or translation. Higher amounts of Cavin3 were observed in V protein-overexpressing cells than in control cells in lipid raft microdomains. Our data collectively suggest that hPIV-2 V protein binds to and stabilizes Cavin3, which in turn facilitates assembly and budding of hPIV-2 in lipid raft microdomains.
Keywords: Cavin3; V protein; caveolae; human parainfluenza virus type 2; lipid raft.
Copyright © 2020 Ohta, Matsumoto and Nishio.
Figures


References
LinkOut - more resources
Full Text Sources

