Linking Comparative Genomics of Nine Potato-Associated Pseudomonas Isolates With Their Differing Biocontrol Potential Against Late Blight
- PMID: 32425922
- PMCID: PMC7204214
- DOI: 10.3389/fmicb.2020.00857
Linking Comparative Genomics of Nine Potato-Associated Pseudomonas Isolates With Their Differing Biocontrol Potential Against Late Blight
Abstract
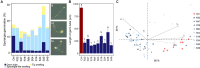
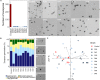
For plants, the advantages of associating with beneficial bacteria include plant growth promotion, reduction of abiotic and biotic stresses and enhanced protection against various pests and diseases. Beneficial bacteria rightly equipped for successful plant colonization and showing antagonistic activity toward plant pathogens seem to be actively recruited by plants. To gain more insights into the genetic determinants responsible for plant colonization and antagonistic activities, we first sequenced and de novo assembled the complete genomes of nine Pseudomonas strains that had exhibited varying antagonistic potential against the notorious oomycete Phytophthora infestans, placed them into the phylogenomic context of known Pseudomonas biocontrol strains and carried out a comparative genomic analysis to define core, accessory (i.e., genes found in two or more, but not all strains) and unique genes. Next, we assessed the colonizing abilities of these strains and used bioassays to characterize their inhibitory effects against different stages of P. infestans' lifecycle. The phenotype data were then correlated with genotype information, assessing over three hundred genes encoding known factors for plant colonization and antimicrobial activity as well as secondary metabolite biosynthesis clusters predicted by antiSMASH. All strains harbored genes required for successful plant colonization but also distinct arsenals of antimicrobial compounds. We identified genes coding for phenazine, hydrogen cyanide, 2-hexyl, 5-propyl resorcinol and pyrrolnitrin synthesis, as well as various siderophores, pyocins and type VI secretion systems. Additionally, the comparative genomic analysis revealed about a hundred accessory genes putatively involved in anti-Phytophthora activity, including a type II secretion system (T2SS), several peptidases and a toxin. Transcriptomic studies and mutagenesis are needed to further investigate the putative involvement of the novel candidate genes and to identify the various mechanisms involved in the inhibition of P. infestans by different Pseudomonas strains.
Keywords: Pseudomonas; beneficial microorganisms; biocontrol; comparative genomics; de novo genome assembly; genotype-phenotype correlation; phyllosphere; rhizosphere.
Copyright © 2020 De Vrieze, Varadarajan, Schneeberger, Bailly, Rohr, Ahrens and Weisskopf.
Figures









References
-
- Acebo-Guerrero Y., Hernández-Rodríguez A., Vandeputte O., Miguélez-Sierra Y., Heydrich-Pérez M., Ye L., et al. (2015). Characterization of Pseudomonas chlororaphis from Theobroma cacao L. rhizosphere with antagonistic activity against Phytophthora palmivora (Butler). J. Appl. Microbiol. 119 1112–1126. 10.1111/jam.12910 - DOI - PubMed
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Andreote F. D., da Roch U. N., Araujo W. L., Azevedo J. L., Van Overbeek L. S. (2010). Effect of bacterial inoculation, plant genotype and developmental stage on root-associated and endophytic bacterial communities in potato (Solanum tuberosum). Antonie Van Leeuwenhoek 97 389–399. 10.1007/s10482-010-9421-9 - DOI - PMC - PubMed
-
- Arseneault T., Goyer C., Filion M. (2016). Biocontrol of potato common scab is associated with high Pseudomonas fluorescens LBUM223 Populations and Phenazine-1-Carboxylic acid biosynthetic transcript accumulation in the potato geocaulosphere. Phytopathology 106 963–970. 10.1094/PHYTO-01-16-0019-R - DOI - PubMed
LinkOut - more resources
Full Text Sources

