AID Overlapping and Polη Hotspots Are Key Features of Evolutionary Variation Within the Human Antibody Heavy Chain (IGHV) Genes
- PMID: 32425948
- PMCID: PMC7204545
- DOI: 10.3389/fimmu.2020.00788
AID Overlapping and Polη Hotspots Are Key Features of Evolutionary Variation Within the Human Antibody Heavy Chain (IGHV) Genes
Abstract
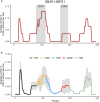
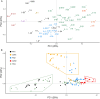
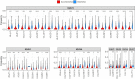
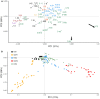
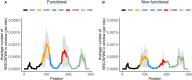
Somatic hypermutation (SHM) of the immunoglobulin variable (IgV) loci is a key process in antibody affinity maturation. The enzyme activation-induced deaminase (AID), initiates SHM by creating C → U mismatches on single-stranded DNA (ssDNA). AID has preferential hotspot motif targets in the context of WRC/GYW (W = A/T, R = A/G, Y = C/T) and particularly at WGCW overlapping hotspots where hotspots appear opposite each other on both strands. Subsequent recruitment of the low-fidelity DNA repair enzyme, Polymerase eta (Polη), during mismatch repair, creates additional mutations at WA/TW sites. Although there are more than 50 functional immunoglobulin heavy chain variable (IGHV) segments in humans, the fundamental differences between these genes and their ability to respond to all possible foreign antigens is still poorly understood. To better understand this, we generated profiles of WGCW hotspots in each of the human IGHV genes and found the expected high frequency in complementarity determining regions (CDRs) that encode the antigen binding sites but also an unexpectedly high frequency of WGCW in certain framework (FW) sub-regions. Principal Components Analysis (PCA) of these overlapping AID hotspot profiles revealed that one major difference between IGHV families is the presence or absence of WGCW in a sub-region of FW3 sometimes referred to as "CDR4." Further differences between members of each family (e.g., IGHV1) are primarily determined by their WGCW densities in CDR1. We previously suggested that the co-localization of AID overlapping and Polη hotspots was associated with high mutability of certain IGHV sub-regions, such as the CDRs. To evaluate the importance of this feature, we extended the WGCW profiles, combining them with local densities of Polη (WA) hotspots, thus describing the co-localization of both types of hotspots across all IGHV genes. We also verified that co-localization is associated with higher mutability. PCA of the co-localization profiles showed CDR1 and CDR2 as being the main contributors to variance among IGHV genes, consistent with the importance of these sub-regions in antigen binding. Our results suggest that AID overlapping (WGCW) hotspots alone or in conjunction with Polη (WA/TW) hotspots are key features of evolutionary variation between IGHV genes.
Keywords: B cell receptor (BCR); activation induced deaminase (AID); computational immunology; dimensionality reduction; immunoglobulin heavy chain; somatic hypermutation (SHM); unsupervised learning.
Copyright © 2020 Tang, Bagnara, Chiorazzi, Scharff and MacCarthy.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

