Integrating NMR and simulations reveals motions in the UUCG tetraloop
- PMID: 32427326
- PMCID: PMC7293013
- DOI: 10.1093/nar/gkaa399
Integrating NMR and simulations reveals motions in the UUCG tetraloop
Abstract
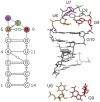
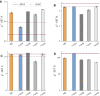
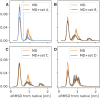
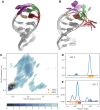
We provide an atomic-level description of the structure and dynamics of the UUCG RNA stem-loop by combining molecular dynamics simulations with experimental data. The integration of simulations with exact nuclear Overhauser enhancements data allowed us to characterize two distinct states of this molecule. The most stable conformation corresponds to the consensus three-dimensional structure. The second state is characterized by the absence of the peculiar non-Watson-Crick interactions in the loop region. By using machine learning techniques we identify a set of experimental measurements that are most sensitive to the presence of non-native states. We find that although our MD ensemble, as well as the consensus UUCG tetraloop structures, are in good agreement with experiments, there are remaining discrepancies. Together, our results show that (i) the MD simulation overstabilize a non-native loop conformation, (ii) eNOE data support its presence with a population of ≈10% and (iii) the structural interpretation of experimental data for dynamic RNAs is highly complex, even for a simple model system such as the UUCG tetraloop.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Cheong C., Varani G., Tinoco I. Jr. Solution structure of an unusually stable RNA hairpin, 5GGAC (UUCG) GUCC. Nature. 1990; 346:680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

