NF90 modulates processing of a subset of human pri-miRNAs
- PMID: 32427329
- PMCID: PMC7337520
- DOI: 10.1093/nar/gkaa386
NF90 modulates processing of a subset of human pri-miRNAs
Abstract
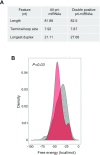
MicroRNAs (miRNAs) are predicted to regulate the expression of >60% of mammalian genes and play fundamental roles in most biological processes. Deregulation of miRNA expression is a hallmark of most cancers and further investigation of mechanisms controlling miRNA biogenesis is needed. The double stranded RNA-binding protein, NF90 has been shown to act as a competitor of Microprocessor for a limited number of primary miRNAs (pri-miRNAs). Here, we show that NF90 has a more widespread effect on pri-miRNA biogenesis than previously thought. Genome-wide approaches revealed that NF90 is associated with the stem region of 38 pri-miRNAs, in a manner that is largely exclusive of Microprocessor. Following loss of NF90, 22 NF90-bound pri-miRNAs showed increased abundance of mature miRNA products. NF90-targeted pri-miRNAs are highly stable, having a lower free energy and fewer mismatches compared to all pri-miRNAs. Mutations leading to less stable structures reduced NF90 binding while increasing pri-miRNA stability led to acquisition of NF90 association, as determined by RNA electrophoretic mobility shift assay (EMSA). NF90-bound and downregulated pri-miRNAs are embedded in introns of host genes and expression of several host genes is concomitantly reduced. These data suggest that NF90 controls the processing of a subset of highly stable, intronic miRNAs.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

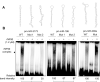
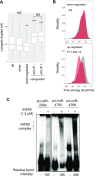
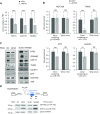
References
-
- Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J.. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004; 432:231–235. - PubMed
-
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R.. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004; 432:235–240. - PubMed
-
- Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

