Targeting the innate immunoreceptor RIG-I overcomes melanoma-intrinsic resistance to T cell immunotherapy
- PMID: 32427578
- PMCID: PMC7410049
- DOI: 10.1172/JCI131572
Targeting the innate immunoreceptor RIG-I overcomes melanoma-intrinsic resistance to T cell immunotherapy
Abstract
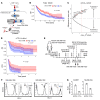
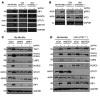
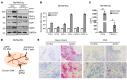
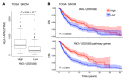
Understanding tumor resistance to T cell immunotherapies is critical to improve patient outcomes. Our study revealed a role for transcriptional suppression of the tumor-intrinsic HLA class I (HLA-I) antigen processing and presentation machinery (APM) in therapy resistance. Low HLA-I APM mRNA levels in melanoma metastases before immune checkpoint blockade (ICB) correlated with nonresponsiveness to therapy and poor clinical outcome. Patient-derived melanoma cells with silenced HLA-I APM escaped recognition by autologous CD8+ T cells. However, targeted activation of the innate immunoreceptor RIG-I initiated de novo HLA-I APM transcription, thereby overcoming T cell resistance. Antigen presentation was restored in interferon-sensitive (IFN-sensitive) but also immunoedited IFN-resistant melanoma models through RIG-I-dependent stimulation of an IFN-independent salvage pathway involving IRF1 and IRF3. Likewise, enhanced HLA-I APM expression was detected in RIG-Ihi (DDX58hi) melanoma biopsies, correlating with improved patient survival. Induction of HLA-I APM by RIG-I synergized with antibodies blocking PD-1 and TIGIT inhibitory checkpoints in boosting the antitumor T cell activity of ICB nonresponders. Overall, the herein-identified IFN-independent effect of RIG-I on tumor antigen presentation and T cell recognition proposes innate immunoreceptor targeting as a strategy to overcome intrinsic T cell resistance of IFN-sensitive and IFN-resistant melanomas and improve clinical outcomes in immunotherapy.
Keywords: Antigen presentation; Immunology; Melanoma; Oncology.
Conflict of interest statement
Figures




Comment in
-
Innate RIG-I signaling restores antigen presentation in tumors and overcomes T cell resistance.Cell Stress. 2021 Jan 18;5(2):26-28. doi: 10.15698/cst2021.02.242. Cell Stress. 2021. PMID: 33554047 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

