Evaluating White Matter Lesion Segmentations with Refined Sørensen-Dice Analysis
- PMID: 32427874
- PMCID: PMC7237671
- DOI: 10.1038/s41598-020-64803-w
Evaluating White Matter Lesion Segmentations with Refined Sørensen-Dice Analysis
Abstract
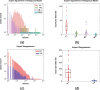
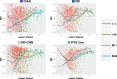
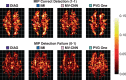
The Sørensen-Dice index (SDI) is a widely used measure for evaluating medical image segmentation algorithms. It offers a standardized measure of segmentation accuracy which has proven useful. However, it offers diminishing insight when the number of objects is unknown, such as in white matter lesion segmentation of multiple sclerosis (MS) patients. We present a refinement for finer grained parsing of SDI results in situations where the number of objects is unknown. We explore these ideas with two case studies showing what can be learned from our two presented studies. Our first study explores an inter-rater comparison, showing that smaller lesions cannot be reliably identified. In our second case study, we demonstrate fusing multiple MS lesion segmentation algorithms based on the insights into the algorithms provided by our analysis to generate a segmentation that exhibits improved performance. This work demonstrates the wealth of information that can be learned from refined analysis of medical image segmentations.
Conflict of interest statement
All undersigned authors certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript. A.C., S.R., A.D., J.C.R., A.J., T.A., O.M., H.H., M.G., B.P., A.B., H.G., D.L.P., C.M.C., W.R.G.R. and I.O. The following authors have declarations: – PAC has received personal consulting fees for serving on SABs for Biogen and Disarm Therapeutics. He is PI on grants to JHU from Biogen, Novartis, Sanofi, Annexon, and MedImmune. – JLP is PI on grants to JHU from Biogen. – RTS has received personal consulting fees from Genentech/Roche.
Figures











References
-
- Zheng, K. Content-based image retrieval for medical image. In 2015 11th International Conference on Computational Intelligence and Security (CIS), 219–222 (2015).
-
- Juang, R. R., Levchenko, A. & Burlina, P. Tracking cell motion using GM-PHD. In 6thInternational Symposium on Biomedical Imaging (ISBI 2009), 1154–1157 (2009).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

