Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability
- PMID: 32429325
- PMCID: PMC7288346
- DOI: 10.3390/genes11050556
Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability
Abstract
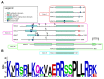
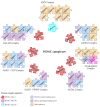
Histone deacetylases (HDACs) are evolutionary conserved enzymes which operate by removing acetyl groups from histones and other protein regulatory factors, with functional consequences on chromatin remodeling and gene expression profiles. We provide here a review on the recent knowledge accrued on the zinc-dependent HDAC protein family across different species, tissues, and human pathologies, specifically focusing on the role of HDAC inhibitors as anti-cancer agents. We will investigate the chemical specificity of different HDACs and discuss their role in the human interactome as members of chromatin-binding and regulatory complexes.
Keywords: HDAC; HDAC inhibitors; HDACi; cancer; chromatin; epigenetics; epigenomics; gene networks; histone deacetylases; phylogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

