Pathway-Specific Genome Editing of PI3K/mTOR Tumor Suppressor Genes Reveals that PTEN Loss Contributes to Cetuximab Resistance in Head and Neck Cancer
- PMID: 32430488
- PMCID: PMC7357849
- DOI: 10.1158/1535-7163.MCT-19-1036
Pathway-Specific Genome Editing of PI3K/mTOR Tumor Suppressor Genes Reveals that PTEN Loss Contributes to Cetuximab Resistance in Head and Neck Cancer
Abstract
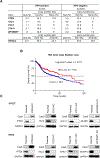
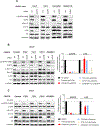
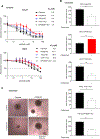
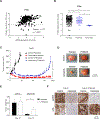
Cetuximab, an mAb targeting EGFR, is a standard of care for the treatment for locally advanced or metastatic head and neck squamous cell carcinoma (HNSCC). However, despite overexpression of EGFR in more than 90% of HNSCC lesions, most patients with HNSCC fail to respond to cetuximab treatment. In addition, there are no available biomarkers to predict sensitivity or resistance to cetuximab in the clinic. Here, we sought to advance precision medicine approaches for HNSCC by identifying PI3K/mTOR signaling network-specific cetuximab resistance mechanisms. We first analyzed the frequency of genomic alterations in genes involved in the PI3K/mTOR signaling circuitry in the HNSCC TCGA dataset. Experimentally, we took advantage of CRISPR/Cas9 genome editing approaches to systematically explore the contribution of genomic alterations in each tumor suppressor gene (TSG) controlling the PI3K-mTOR pathway to cetuximab resistance in HNSCC cases that do not exhibit PIK3CA mutations. Remarkably, we found that many HNSCC cases exhibit pathway-specific gene copy number loss of multiple TSGs that normally restrain PI3K/mTOR signaling. Among them, we found that both engineered and endogenous PTEN gene deletions can mediate resistance to cetuximab. Our findings suggest that PTEN gene copy number loss, which is highly prevalent in HNSCC, may result in sustained PI3K/mTOR signaling independent of EGFR, thereby representing a promising mechanistic biomarker predictive of cetuximab resistance in this cancer type. Further prospective studies are needed to investigate the impact of PTEN loss on cetuximab efficacy in the clinic.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin [Internet]. 2019;69:7–34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30620402 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

