A comparative look at structural variation among RC-LH1 'Core' complexes present in anoxygenic phototrophic bacteria
- PMID: 32430765
- PMCID: PMC7423801
- DOI: 10.1007/s11120-020-00758-3
A comparative look at structural variation among RC-LH1 'Core' complexes present in anoxygenic phototrophic bacteria
Abstract
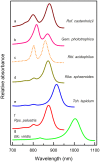
All purple photosynthetic bacteria contain RC-LH1 'Core' complexes. The structure of this complex from Rhodobacter sphaeroides, Rhodopseudomonas palustris and Thermochromatium tepidum has been solved using X-ray crystallography. Recently, the application of single particle cryo-EM has revolutionised structural biology and the structure of the RC-LH1 'Core' complex from Blastochloris viridis has been solved using this technique, as well as the complex from the non-purple Chloroflexi species, Roseiflexus castenholzii. It is apparent that these structures are variations on a theme, although with a greater degree of structural diversity within them than previously thought. Furthermore, it has recently been discovered that the only phototrophic representative from the phylum Gemmatimonadetes, Gemmatimonas phototrophica, also contains a RC-LH1 'Core' complex. At present only a low-resolution EM-projection map exists but this shows that the Gemmatimonas phototrophica complex contains a double LH1 ring. This short review compares these different structures and looks at the functional significance of these variations from two main standpoints: energy transfer and quinone exchange.
Keywords: Anoxygenic phototrophs; Light harvesting; Purple photosynthetic bacteria; RC–LH1; Reaction centres; Structures.
Conflict of interest statement
There are no conflicts of interest.
Figures



References
-
- Barz WP, Francia F, Venturoli G, Melandri BA, Vermeglio A, Oesterhelt D. Role of PufX protein in photosynthetic growth of Rhodobacter sphaeroides 1 PufX is required for efficient light-driven electron transfer and photophosphorylation under anaerobic conditions. Biochemistry. 1995;34:15235–15247. doi: 10.1021/bi00046a032. - DOI - PubMed
-
- Barz WP, Vermeglio A, Francia F, Venturoli G, Melandri BA, Oesterhelt D. Role of the PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 2. PufX is required for efficient ubiquinone/ubiquinol exchange between the reaction center QB site and the cytochrome bc1 complex. Biochemistry. 1995;34:15248–15258. doi: 10.1021/bi00046a033. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

